Background
Marbling is the most economically important trait in beef cattle industry of Korea. Generally, marbling means the amount and distribution of intramuscular fat in a cross section of musculus longissimus muscle [1]. In particular, eating quality traits such as taste, juiciness and tenderness of meat are influenced by the amount of intramuscular fat [2]. High levels of marbling improve the palatability and acceptability of beef by affecting the taste and tenderness of the meat [3, 4]. Therefore, the challenge to the beef cattle industry in Korea is the production of meat with higher marbling score. Knowledge on the genetic background of fat tissue accumulation and better understanding of the molecular mechanism of marbling are very important in beef production. To identify an informative gene or DNA marker for meat quality traits, candidate genes can be selected on the basis of the function of the encoded protein in physiological processes controlling energy homeostasis.
Recently, the fat mass and obesity associated (FTO) gene have been shown to have a relatively large effect on body mass index (BMI) and obesity-related traits in various human populations, suggesting that FTO associated with the development of fat tissue and adiposity [5–9]. Some studies also suggested that the FTO may play a key role in the regulation of energy homeostasis and associated with increased lipolytic activity in adipose tissue [9, 10]. In livestock species, polymorphic variations in the FTO gene are associated with fatness-related traits such as intramuscular fat deposition and backfat thickness [11–15]. In addition, the bovine FTO gene is located near the QTL region affecting meat quality traits on BTA18. Therefore, the FTO gene is considered as a positional and functional candidate gene for meat quality in beef cattle. However, most studies have so far been published for the pig. Moreover, the association analysis between single nucleotide polymorphisms (SNPs) within exons of the FTO gene and meat quality traits has not been reported in Hanwoo (Korean cattle). The objectives of our study were to identify new SNPs in the exonic regions of FTO gene and to evaluate the association of these SNPs with meat quality traits in Hanwoo.
Methods
A total of 300 steers, which were registered in a national database of cattle and guided with standardized breeding programs provided by the Hoengseong Hanwoo population in Korea, were used to genotype and collect carcass traits. To confirm genetic inheritance of the identified SNPs, 51 Hanwoo animals, which were used for the evaluation of performance ability for proven sires in the national progeny testing programs, were genotyped. The carcass data analyzed in the current study included marbling score (MS), meat color (MC), fat color (FC), meat texture (MT), meat maturity (MA), backfat thickness (BF), Longissimus dorsi muscle area (LMA), and carcass weight (CW). The means carcass traits values were 5.98 ± 1.73 for MS, 4.91 ± 0.37 for MC, 3.00 ± 0.10 for FC, 1.09 ± 0.28 for MT, 2.03 ± 0.19 for MA, 11.99 ± 4.01 mm for BF, 90.05 ± 9.57 cm2 for LMA and 427.11 ± 46.96 kg for CW.
Meat samples were collected from 13th thoracic rib to the first lumbar vertebrae of the steers within 24 hr of slaughter and evaluated according to the Animal Product Grading System of Korea. Genomic DNA was extracted from tail root-hair by using a NaCl precipitation protocol [16] with a slight modification. The DNA sample was suspended in TE buffer (10 mM Tris–HCl, pH 7.4; 1 mM EDTA) and stored at -20°C until analysis.
The bovine FTO gene is mapped to chromosome 18, which includes nine exons coding 505 amino acids (Figure 1). To identify polymorphisms within the coding region of the Hanwoo FTO gene, the nine pairs of primers (Table 1) were designed to amplify the all exon regions based on the genomic sequence of the bovine FTO gene from NCBI GenBank reference sequences (Genomic sequence: AC_000175.1 chromosome 18 reference Bos taurus UMD 3.1 primary assembly) using a web-based software Primer 3.0 program (http://frodo.wi.mit.edu/primer3). To determine SNP identification, pooled DNA samples from the sixty unrelated animals were amplified by PCR using the each primer pair. The PCR amplification was performed in a DNA thermal cycler (Perkin Elmer Cetus, Norwalk, CT). The PCR reaction was performed in a 20 μL reaction mixture containing 0.1 μM of each primer, 1.5 mM MgCl2, 250 μM of each dNTP and 1 unit of Taq DNA polymerase, 10 X reaction buffer and 50 ng of pooled DNA as template. The PCR conditions were at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, annealing at 55°C to 57°C for each primer for 20 s and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR products were verified by 2% agarose gel electrophoresis and purified with Wizard Prep PCR purification kit (SolGent, Korea). The purified PCR amplicons were directly sequenced in both directions using BigDye™ Terminator V3.1 Cycle Sequencing Kit in an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. GENESCAN 3.7 ANALYSIS software (Applied Biosystems, USA) was used to assemble the sequences and to identify polymorphisms.
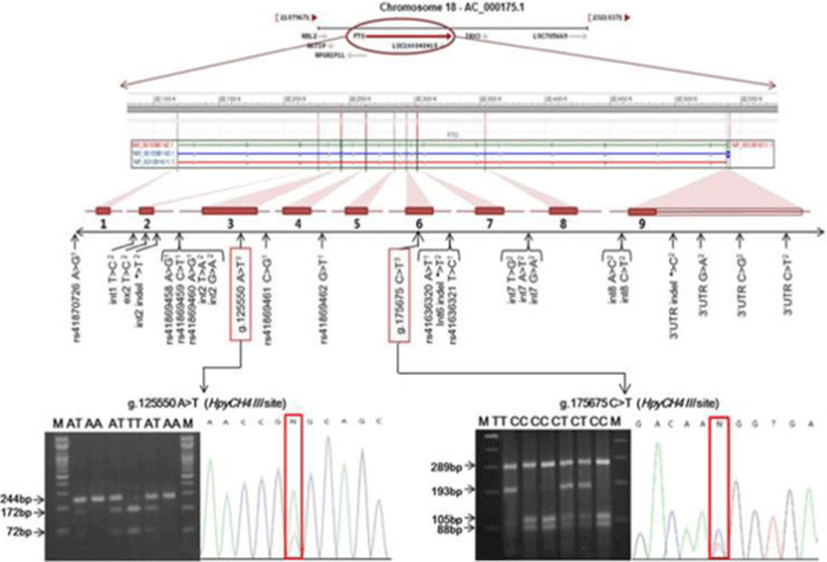
Two SNPs, g.125550A > T and g.175675C > T, newly identified in the exons 3 and 6 of the Hanwoo FTO gene respectively, were genotyped using PCR-RFLP method. PCR primers used for PCR-RFLP analysis were 5'-TTCCTCAAGCTCAACAGCTACC-3' and 5'-ACGGTTCCTCTTTCAGGTATGG-3' for the g.125550A > T SNP, and 5'-AGAGTCAGTTCTAGGTGCTGTGGT-3' and 5'-CCACAGTTCTCAGAAGCCCTTA-3' for the g.175675C > T SNP. PCR amplifications were carried out as for the SNP detection protocol. For the RFLP analysis, amplified fragments were digested with restriction enzyme HpyCH4III for g.125550A > T and g.175675C > T at 37°C for 3 h, respectively. The digested DNA fragments were separated on 2% agarose gel by electrophoresis with 1 X TBE buffer. The gels were stained with ethidium bromide (EtBr) and the fragments were visualized using a UV transilluminator (Ultra Rum Inc, USA). To define each genotype according to band patterns, the PCR products of different RFLP type corresponding to each genotype were sequenced and analyzed for nucleotide changes.
Allele and genotype frequencies of SNPs and Hardy-Weinberg equilibrium were estimated and tested using PROC ALLELE (SAS Inst. Inc., Cary, NC, USA). Associations between SNP genotypes and phenotypes of carcass traits were analyzed with the following liner mixed model using the MIXED procedure of SAS (SAS Inst. Inc., Cary, NC, USA). The data were analyzed according to the following model: Yijk = μ + Gi + Sj + Pk + βAl + eijkl, Where Yijk =the value of carcass traits; μ = the overall mean for each trait; G = fixed effect of single SNP marker genotype; S = random effect of sire, P = fixed effect of parity; A = fixed effect of age as a covariate; and eijkl = random residual effect. The Bonferroni correction for multiple testing was performed to remove any false positives. The additive and dominance genetic effects were also estimated using REG procedure of SAS according to Falconer and Mackay [18]. This research was followed by internationally recongnized guidelines (Institutional Animal Care and Use Committee, IACUC) for animal experiment.
Results
In this study, we sequenced all exon regions of the bovine FTO gene and newly identified two exonic SNPs in Hanwoo steers (Figure 1). An A to T transition (g.125550A > T SNP) was located in exon 3 and a C to T substitution (g.175675C > T SNP) was located in exon 6 (GenBank accession no. AC_000175). Genotyping of the two novel SNPs in the coding region of the bovine FTO gene was performed by a PCR-RFLP method in Hanwoo population. The allele and genotype frequencies in the two novel SNPs of the FTO gene are shown in Table 2. The genotypic frequencies were as follows: 47.0% AA, 44.3% AT and 8.7% TT for the g.125550A > T SNP; 52.0% CC, 37.3% CT and 10.7% TT for the g.175675C > T SNP. The observed genotype distributions were in good agreement with those expected according to the Hardy-Weinberg equilibrium in this population. Overall average of heterozygosity (He) and polymorphic information contents (PIC) for two SNP markers were calculated to 0.576 and 0.486, respectively. The results of the association analysis for the FTO gene SNP markers with various carcass traits are presented in Table 3. The g.125550A > T SNP genotype was significantly associated with effects on MS. Animals with the AA or TT homozygous genotype had a significantly higher MS (p < 0.001) than the animals with AT heterozygous genotype and this was significant after Bonferroni correction of the significance threshold (p = 0.003). Dominance effect was also observed for the marbling score (P < 0.05) with higher marbling score of homozygous animals. However, no significant association was detected between the g.175675C > T SNP genotype and carcass traits measured in this study.
He: Heterozygosity, PIC: Polymorphic Information Content, HWE: Hardy-Weinberg Equilibrium.
1 MS, marbling score; MC, meat color; FC, fat color; MT, meat texture; MA, meat maturity; BF, backfat thickness; LMA, M. Longissimus dorsi muscle area; CW, carcass weight.
***P <0.001.
a,bWithin a row, means with different superscripted letters are different (P <0.01).
Discussion
The development of functional genomics has revealed a large number of genes related with fat accumulation and lipid metabolism. Several QTLs for carcass traits and a large number of potential candidate genes based on a known relationship with physiological or biochemical processes and carcass traits have been reported in beef cattle [19–21]. However, a limited number of genetic markers have been recognized for carcass and meat quality traits in beef cattle and these markers explain a relatively small proportion of the genetic variation for a limited number of traits [22]. Fatness related traits such as marbling are classified as a quantitative trait with a high contribution of genetic variation [23]. Thus, identification of DNA polymorphism associated with (or responsible for) fatness traits may be useful for marker assisted selection.
Many regulatory factor genes are involved in the formation of marbling [24], and increased knowledge about the relationship between these genes and the fat accumulation and lipid metabolism is of utmost importance for the improvement of meat quality in beef cattle. The FTO gene is a new candidate gene related to the development of fat tissue and obesity. This gene encodes 2-oxoglutarate-dependent oxygenases, which are involved in various processes, including DNA repair, fatty acid metabolism and posttranslational modifications and is highly expressed in the hypothalamic pituitary adrenal axis, which plays a key role in the control of energy balance [25]. Therefore, the polymorphic variation in this gene may play a causal role in the regulation of energy homeostasis or in the development of fat tissue [5, 26]. In addition, the FTO is a transcriptional co-activator, which facilitates transcription from unmethylated and methylation-inhibited gene promoters and enhances C/EBPs binding to DNA, and that it may play a role in the regulation of adiposity [15, 27]. These findings suggest close link between FTO and fat deposition and lipid metabolism. In human, genetic variation of this gene was extensively studied and had confirmed that a strong and highly significant association with fatness-related traits such as fat mass, obesity and body mass index in different populations [5–7]. This gene has also become one of the candidate genes for detecting polymorphism associated with the fat deposition in livestock species. Several studies on pig have shown the association between the polymorphism of the FTO gene and intramuscular fat (IMF) [11, 13, 28]. A significant association between g.276 T > G polymorphism of the 3'-untranslated region and intramuscular fat deposition was reported by Fontanesi et al. [28] in the Italian White Duroc breed. Fan et al. [12] also suggested significant association of the g.-1191A > G SNP in 5′ regulatory region of the porcine FTO gene and intramuscular fat content. These results provided that the porcine FTO gene might play an important function in intramuscular fat and not directly in subcutaneous or abdominal fat deposition [13]. Recently, some previous studies have reported the bovine FTO gene associated with carcass traits in beef cattle breeds. Wei et al. [14] have reported that g.1071C > T SNP in exon 5 within the bovine FTO gene was associated with backfat thinckness and longissmus muscle area traits in five Chinese native cattle breeds. In this study, however, the effect of genotypes of the two novel SNPs was not statistically significant for backfat thickness and longissmus muscle area. This might be supported by the fact that Hanwoo breed exhibits very low or negative genetic correlation between marbling and backfat thickness [29, 30]. Also, Jevšinek Skok et al. [31] have reported that the T > C SNP in exon 2 of the FTO gene showed a significant effect on growth and carcass traits such as live weight at slaughter, carcass weight and lean weight in Slovenian Simmental population. However, the associations between the SNPs of the FTO gene and intramuscular fat have not been reported in beef cattle. To the knowledge of the authors, the present study is the first report on association between exonic SNP marker within coding regions of the FTO gene and fatness-related traits such as marbling score in beef cattle, suggesting a possible effect of the gene on intramuscular fat deposition. The two exonic SNPs identified in this study by resequencing have not been reported previously and are therefore novel. The bovine FTO gene was near to the QTL region for carcass traits [32, 33]. As the A to C substitution in exonic SNP g.125550A > T does not result in an amino acid change due to synonymous mutation, this is unlikely to be the causal mutation for marbling and instead is likely to be genetically linked to nearby QTL or causative mutations that affect marbling score in beef cattle. The synonymous SNPs could affect gene function because they are under evolutionary pressure, alter the structure, function and expression level of proteins, and can affect mRNA splicing, stability and protein structure as well as folding [34].
Conclusion
In conclusion, we identified two novel exonic SNPs of the FTO gene in Hanwoo population and the g.125550A > T SNP genotype showed significant effect on marbling score. These findings suggest that the FTO gene-specific SNP identified in this study may be useful molecular marker for selection to increase the levels of marbling in Hanwoo. This study also will contribute to a better understanding of the molecular mechanisms of marbling in beef cattle. Further studies are will be needed to confirm the associated effect on other population and breeds.
