INTRODUCTION
Innate immunity is the first defense line against various pathogens through sensing pathogens, eliminating them, and activating adaptive immune response [1]. In sensing pathogens, nucleic acids (NAs) that are originated from pathogenic bacteria and viruses are recognized by innate immune receptor signaling, which are mediated by pattern recognition receptors (PRRs) including toll-like receptors (TLRs), retinoic acid inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) [2,3]. Among them, TLR3, TLR7/8, TLR9, and TLR13 of TLRs are known as nucleic acid NA-sensing TLRs. They primarily exist in endosome and respond to double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), single-stranded DNA, and bacterial ribosomal RNA respectively [4,5]. RIG-I, MDA5, and LGP2 are cytosolic NA receptors which detect dsRNA. RIG-I primarily responds to 5’-triphosphorylated blunt-ended RNA or dsRNA produced during RNA virus infections and MDA5 responds to long dsRNA [2]. LGP2 also seems to enhance initial MDA5-RNA interaction [6]. Complex with cognate PRRs and their ligands leads to the engagements of myeloid differentiation primary response 88 (MYD88), Toll/IL-1R homologous region (TIR) domain-containing adapter-inducing interferon-β (TRIF), or mitochondrial antiviral-signaling protein (MAVS). It activates transcription factors (TFs) such as interferon regulatory factor3 (IRF3), IRF7, nuclear factor kappa B (NF-κB), and activating protein 1 (AP-1) (ATF2/JUN) by orchestrating a combination of multi-protein complexes. The TFs induce to express inflammatory cytokines, chemokines and type I interferons [7–12].
Among the NA-sensing TLRs, chickens have obvious orthologues of TLR3 and TLR7 while TLR8 has been disrupted by the insertion of a large CR1 repeat [13]. TLR9 and TLR13 were also absent [8,14]. In addition, TLR15 and TLR21 uniquely existed in chickens compared to human and mouse [15,16]. Chicken TLR21 has recently been shown to recognize CpG motifs, suggesting a functional homologue to mammalian TLR9 [17] whereas an virus-related agonist for TLR15 remains unknown [18,19]. MDA5 and LGP2 are also present in chicken genome and their function seems to be similar to mammals whereas RIG-I is obviously absent [20,21]. It has been suggested that the lack of RIG-I caused a susceptibility for zoonotic RNA virus such as avian influenza in chickens [22]. Even if immune responses to NA have been comparatively well characterized in chickens, the precise mechanism remains to be elucidated.
Polyinosinic:polycytidylic acid (poly[I:C]), viral like dsRNA, has generally been used to mimic NA-sensing responses of the innate immune system. Poly(I:C) is recognized by TLR3 and MDA-5, activate various TFs such as IRFs and NF-κB, and stimulates various cytokines and chemokine, IFNs and costimulatory factors in various species [10,11,23–26]. Poly(I:C) exhibited a toxicity in various tissues and cells [27,28]. Especially, the viability of chicken embryonic fibroblasts (CEFs) reduced to about 80% and below 50% with 1,000 µg/mL of poly(I:C) for 24 h and 72 h respectively and it suggested that poly(I:C) induced apoptosis of CEFs through the activation of caspase-3 and -8 by TNFRSF8 [29]. In addition, DF-1 cells, chicken fibroblast cell line modulated IRF7-related immune signaling pathways responding to poly(I:C) [30]. In this regard, chicken fibroblasts including DF-1 are a useful model to study in vitro immune responses which are stimulated by poly(I:C).
In this study, we examined the expression patterns of innate immune signaling-related genes such as canonical and non-canonical TLRs, the related TFs, cytokines, and immune-related effector molecules in chickens after poly(I:C) treatment. Our results could contribute to understanding the gene expression which is involved in NA-sensing and the related responses in chicken cells.
MATERIALS AND METHODS
DF-1 chicken fibroblast cell lines were obtained from the American Type Culture Collection (Rockville, MD, USA) and maintained in the Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Biowest, Nuaillé, France). DF-1 cells were cultured at 37°C in 5% CO2 incubator. Poly(I:C) was purchased from Invivogen (San Diego, CA, USA) and was stocked according to the manufacturer’s instruction and all poly(I:C) treatment was maintained under the culture condition of DF-1 cells.
Cell viability assays were performed using tetrazolium compound based CellTiter 96® AQueous One Solution Cell Proliferation (MTS) assay (Promega, Madison, WI, USA). MTS assay was then performed according to the manufacturer’s instruction at 24 h after treatment at indicated concentrations of poly(I:C).
RNAs were isolated from DF-1 cells using RNA extraction kit (Invitrogen, CA, USA). For quantitative reverse transcription-polymerase chain reaction (qRT-PCR), 1 µg of total RNA was used for cDNA synthesis with Rever Tra Ace-α- first strand cDNA Synthesis Kit (Toyobo, Osaka, Japan). Sequence-specific primers (Table 1) were designed using the Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). qRT- PCR was performed using the iCycler real-time PCR detection system (Bio-Rad, Hercules, CA, USA) and SYBR Green (Bio-Rad, Hercules, CA, USA). Non-template wells without cDNA were included as negative controls. Each sample was tested in triplicate. The PCR conditions were 95°C for 3 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s, using a melting curve program (increasing temperature from 65°C to 95°C at a rate of 0.5°C per 5 s) and continuous fluorescence measurement. The qRT-PCR data were normalized relative to the expression of GAPDH and calculated using the 2 ∆∆Ct method, where ∆∆Ct = (Ct of the target gene – Ct of GAPDH) treatment – (Ct of the target gene – Ct of GAPDH) control [31].
RESULTS AND DISCUSSION
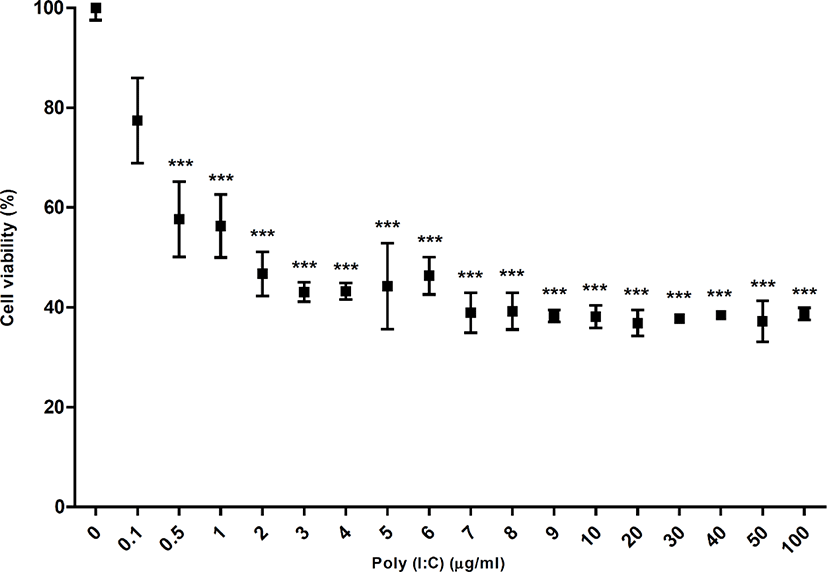
In this study, poly(I:C) treatment with different doses from 0.1 µg/mL to 100 µg/mL for 24 h decreased the viability of DF-1 cells (chicken fibroblasts cell line) by 77.41%, 57.63%, 56.28%, 46.69%, 43.06%, 43.19%, 44.22%, 43.32%, 38.9%, 39.19%, 38.25%, 38.1%, 36.85%, 37.73%, 38.42%, 37.17%, and 38.68% respectively, compared to the non-treated control. The statistical analysis showed significant difference at all the treated concentrations except the concentration of 0.1 µg/mL, compared to the non-treated control and no difference among the cell viabilities from 0.5 µg/mL to 100 µg/mL poly(I:C) (p < 0.05) (Fig. 1). These results suggested that poly(I:C) rapidly affected on the cell viability from 0.5 µg/mL and this effect was saturated from 0.5 µg/mL to 100 µg/mL. Thus, we supposed that DF-1 cells could be much more sensitive to poly(I:C) than primary cultured CEFs.
TLR3 and TLR7 are known as NA-sensing TLRs while the function of TLR4 is associated with the recognition of endotoxins molecules, in particular lipopolysaccharide from gram-negative bacteria [13,32]. Recently, the several studies have shown that TLR3, 4, and 7 mediated the responses to the viral-associated PAMPs such as poly(I:C), F protein of Respiratory Syncytial Virus (RSV), and imidazoquinolines, antiviral therapeutic compounds, respectively [33–38]. In addition, it has been reported that selective activation of TLR3/4-IRF3 pathway was associated with potential inhibition of viral replication [39]. TLR15, an avian-specific TLR, has been reported to be induced by salmonella, mycoplasma, and even Marek’s Disease Virus (MDV) [16,18,19,40]; however, the specifically virus-associated agonist was still unknown [41]. Instead of mammalian TLR9 which was missing from the chicken genome, chicken TLR21 acted as a functional homologue to the mammalian TLR9 to recognize CpG [17]. Poly(I:C) and CpG ODN (CpG-motif containing oligodeoxydinucleotide) synergized the expression of pro-inflammatory cytokines and chemokines and the production of nitric oxide in chicken monocytes [42,43].
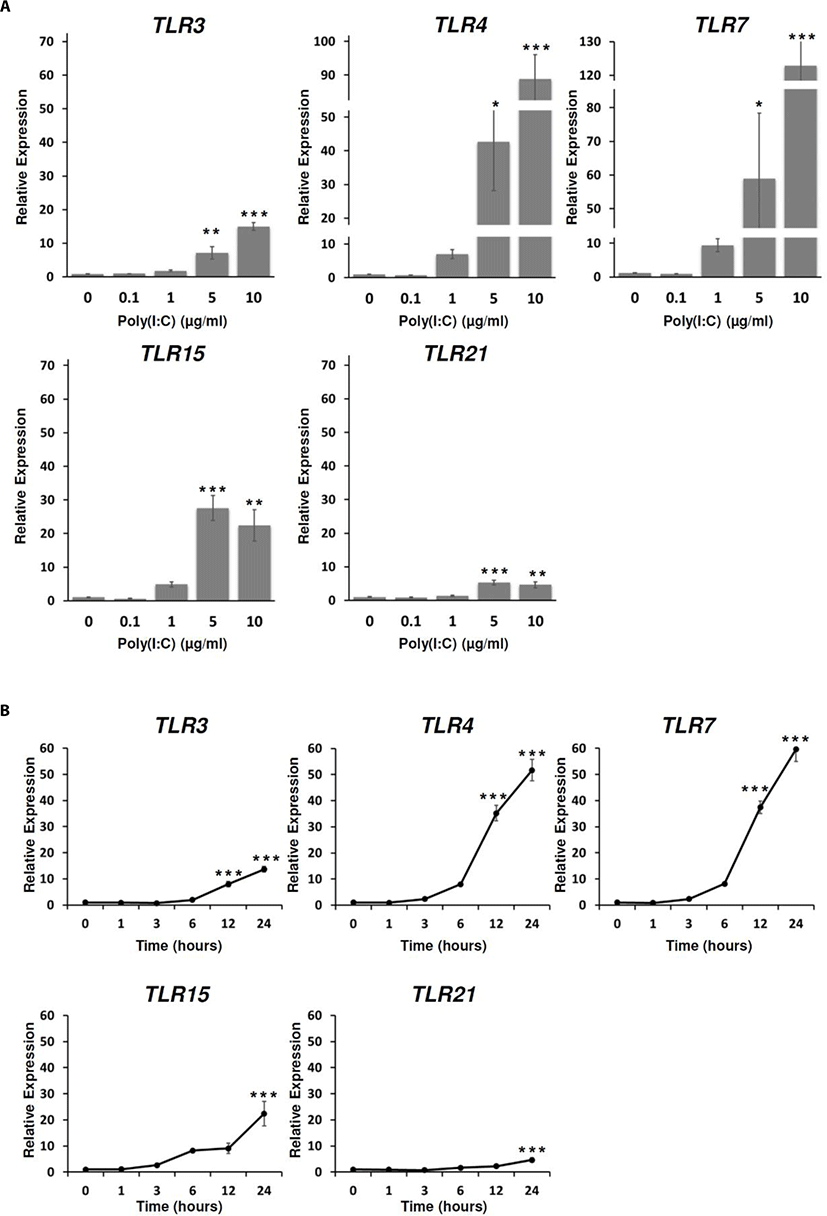
To investigate chicken TLRs expressions in response to poly(I:C) treatment, the expressions of chicken TLRs were analyzed dose and time-dependently. From the analysis, the expressions of TLR3, 4, 7, 15, and 21 were significantly induced at the poly(I:C) concentrations of 5 µg/mL and 10 µg/mL for 24 h (Fig. 2A). In addition, the expression levels of TLR3, 4, 7, 15, and 21 were significantly increased with 10 µg/mL poly(I:C) at 12 h and 24 h after poly(I:C) treatment (Fig. 2B). Therefore, we suggested that poly(I:C) was directly targeted at these TLRs in DF-1 cells to stimulate immune responses.
TLRs which recognize their ligands activated conserved TFs including AP-1, NF-κB, and IRFs through the interplay of complex TLR signaling pathways [44–47]. Among the AP-1 family, JUN that was a target protein of c-Jun N-terminal kinase (JNK) was regarded as a key factor in TLR signaling [47]. Among NF-κB protein complex, NF-κB1 (also known as p50) was known to have DNA binding activity for the promoter region of its target genes [48]. Among IRFs, IRF3 and IRF7 were activated by various ligands, such as poly(I:C), LPS, and virus infection and mainly controlled type-I IFN expression [49]. In mammalian, type I IFNs-mediated signaling pathways were dependent on the stimulus and the responding cell types. TLR signaling pathways associated with type I IFN, TLR3 and TLR4 induced type I IFN production in various cell types in a manner dependent on TIR-domain-containing adaptor protein inducing IFNβ (TRIF) whereas TLR7, TLR8 and TLR9 induced type I IFN production in dendritic cells via a pathway dependent on MYD88. Eventually they can activate some common signaling molecules including TNF receptor-associated factor 3 (TRAF3) and IRF3 and IRF7 [49,50]. Additionally, poly(I:C) treatment increased IRF7 and type-I IFN (IFNA) in DF-1 cells [25].
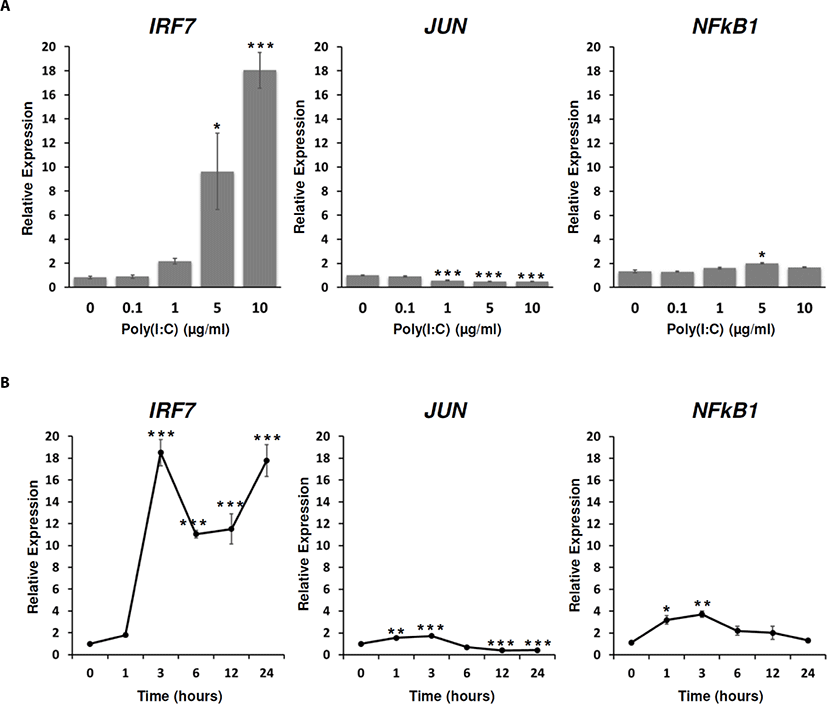
To reveal TFs which are associated with TLR signaling responded to poly(I:C), the expressions of IRF7, JUN, and NF-κB1 were analyzed in DF-1 cells at different doses of poly(I:C) and time points. From the dose-dependent treatment, IRF7 and NF-κB1 expressions were significantly increased at 5 µg/mL and 10 µg/mL and 5 µg/mL of the poly(I:C) treatment for 24 h, respectively. Whereas the expression of JUN was significantly decreased at 1 µg/mL, 5 µg/mL, and 10 µg/mL of poly(I:C) for 24 h (Fig. 3A). When the expressions of IRF7, JUN, and NF-κB1 were analyzed with 10 µg/mL poly(I:C) according to time course, the expression of IRF7 steadily increased from 3 h to 24 h after poly(I:C) treatment. JUN and NF-κB1 expressions were commonly increased from 1 h to 3 h after poly(I:C) treatment, but were decreased from 6 h to 24 h after poly(I:C) treatment (Fig. 3B). These results suggested that TLR3 stimulation by poly(I:C) induced IRF7 transcription, whereas the expressions of JUN and NF-κB1 were gradually decreased and maintained to the ground state although they were rapidly induced within 1 h after the poly(I:C) treatment. Thus, we speculated that poly(I:C) may mainly induce immune-effector genes by IRF7-mediated signaling pathway after the recognition by TLRs such as TLR3, 4, 7, 15, and 21 in 24 h after the treatment while direct or indirect pathways may exist to acutely induce JUN and NF-κB1. The further study is necessary to prove the activation of TLR pathway-mediated TFs.
From TLRs recognizing their ligands, the activated TFs can induce a variety of interferons, cytokines and chemokines [9,39]. During the immune responses, cytokine and chemokine families acted as extracellular molecular regulators which mediated immune cell recruitment and participated in complex intracellular signaling processes [9]. Among them, IL1B belonging to IL1 family and IL10 have been known as a pro-inflammatory and an anti-inflammatory cytokine respectively. These cytokines were induced by viral infections [9,51–53]. IL8, a critical inflammatory chemokine was also upregulated by various viral infection in human epithelial cells [54].
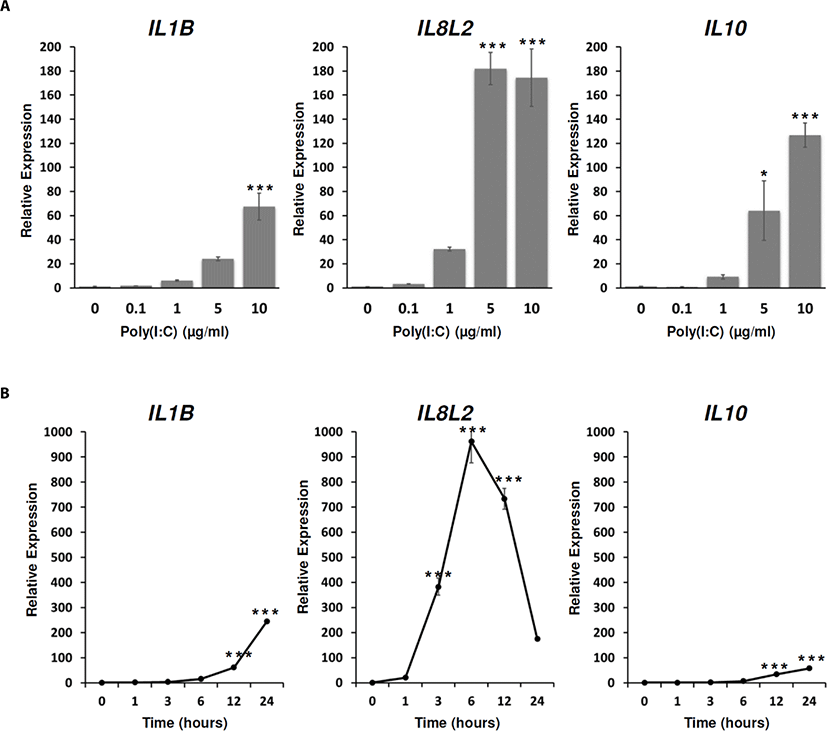
To examine whether the expressions of immune-related effector genes are affected by poly(I:C) treatment, IL1B, IL8L2 (chicken IL8-like 2), and IL10 expressions were analyzed after the poly(I:C) treatment at different dose and time points. From the analysis, the expressions of IL1B,IL8L2, and IL10 were significantly increased by poly(I:C) treatments from 5 µg/mL to 10 µg/mL for 24 h (Fig. 4A). In time-dependent analysis, the expressions of IL1B, and IL10 were significantly increased from 12 h to 24 h after the poly(I:C) treatment (Fig. 4B). Unlike IL1B and IL10, the expression of IL8L2 showed the rapid increase at 3 h after the poly(I:C) treatment and reached to the plateau at 6 h after the poly(I:C) treatment. In addition, it was continuously decreased from 12 h to 24 h after the poly(I:C) treatment compared to the expression of IL8L2 at 6 h after the poly(I:C) treatment although the expressions of IL8L2 at 12 h and 24 h after the poly(I:C) treatment were still higher than the non-treated control (Fig. 4B). This result suggested that the inductions of IL1B,IL8L2, and IL10 in DF-1 cells could be mediated by TLR-signaling pathways. In addition, IL8L2 could more sensitively respond to poly(I:C) and be inhibited by other feedback systems compared to IL1B and IL10.
Conclusively, we suggested the distinct TLR signaling pathways which responded to poly(I:C) in chicken-originated cell line (DF-1) compared to mammalian TLRs for NA-sensing and their signaling pathways. Our results could contribute to understanding NA-sensing and subsequent immune signaling pathways in chicken cells.
