Staphylococcus aureus is a Gram-positive bacteria that is detected in the environment and also existing in normal human microbiome [1]. In addition, this bacterium is a representative bacterial human pathogen and contaminated with food through purulent wounds of humans and animals. Particularly, methicillin-resistant S. aureus (MRSA) has appeared as a significant issue, with important anxieties about public health because they can spread easily through healthy carriers and increase the likelihood of infection by expressing a number of toxic factors, cell wall-related adecines and secreted exoproteins [2]. Therefore, monitoring of S. aureus in a dairy environment with high sensitivity is very important for ensuring milk quality and food safety [3].
| Name | Length (bp) | GC (%) | Depth | CDSs | tRNA | rRNA |
|---|---|---|---|---|---|---|
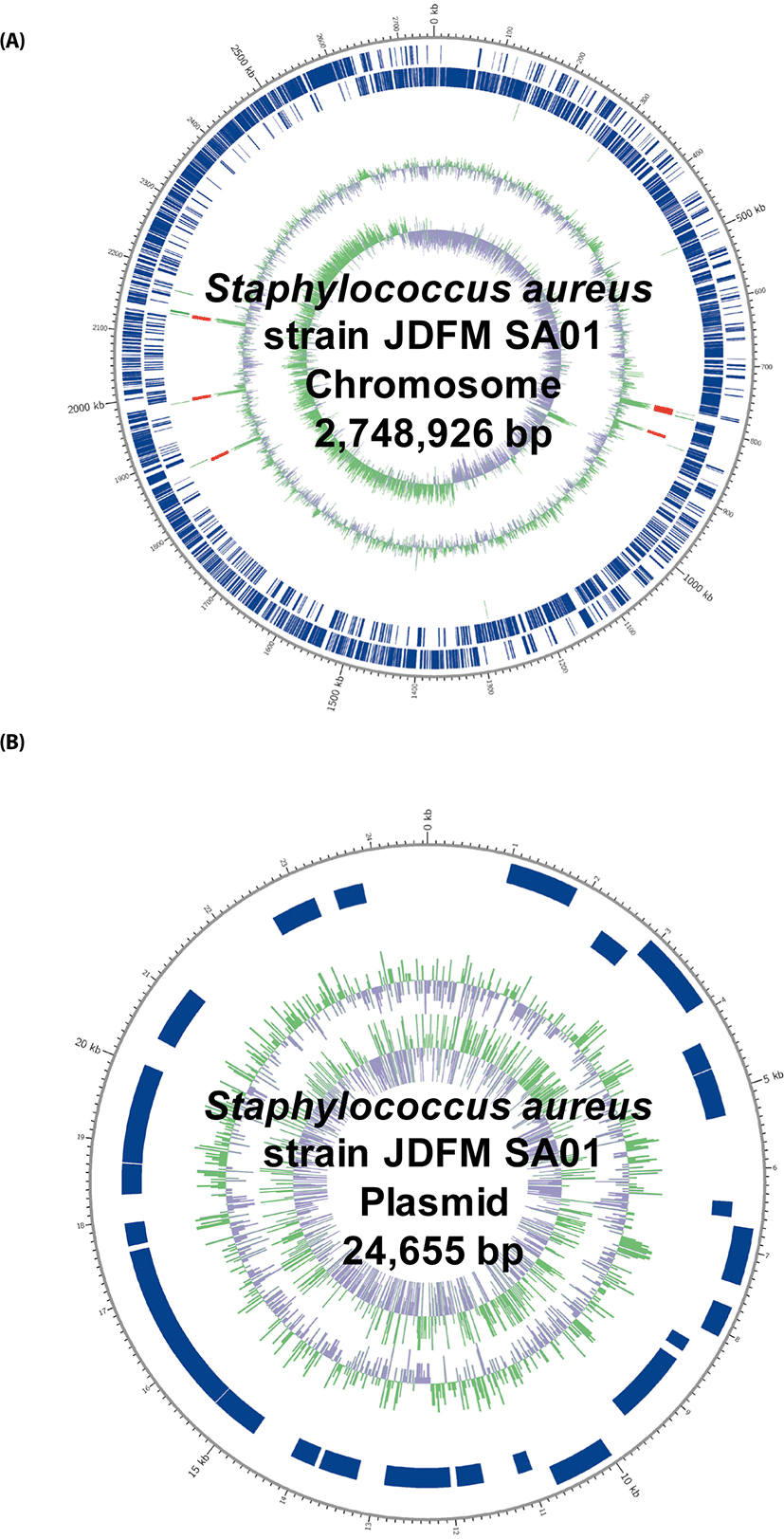
| Chromosome | 2,748,925 | 32.9 | 408 | 2,520 | 61 | 19 |
| Plasmid | 24,655 | 28.7 | 323 | 29 | 0 | 0 |
| Total | 2,773,580 | 32.84 | 407 | 2,549 | 61 | 19 |
In the present study, S. aureus strain JDFM SA01 was isolated from a milk filter in Korean dairy farm (Jeollabuk-do; sampled during May 2017 to May 2018). The sample was added to 10% NaCl-added Tryptic Soy Broth (TSB) and incubated at 37°C for 24 h. The enrichment culture was inoculated onto Baird Parker (Oxoid) agar and cultured at 37°C for 24 h [4]. After incubation, a single colony, designated S. aureus strain JDFM SA01, was selected and routinely maintained on tryptic soy broth at 37°C for sequencing. Total genomic DNA was extracted using the PureHelixTM GenomicDNA prep kit (Nanohelix, Korea), according to the manufacturer’s instructions.
The whole genome of S. aureus strain JDFM SA01 was sequenced by using the Pacbio RS II (Pacific Biosciences, USA) / Illumina HiSeq (151 × 2 bp paired-end sequencing) platforms at Macrogen (Seoul, Korea). Library preparation for Illumina and PacBio sequencing was performed using the NEBNext Ultra DNA library prep kit for Illumina (NE, USA) and the PacBio DNA template prep kit 1.0 (Pacific Biosciences, USA), respectively, according to the manufacturers’ instructions. A total number of 179,260 reads with a mean subread length of 8,498 bases (N50, 12,066 bases) were obtained with PacBio sequencing, and 10,257,096 paired-end reads totaling 1,548,821,496 bp were obtained with Illumina sequencing. De novo assembly was carried out using the Hierarchical Genome Assembly Process v3.0 (HGAP3) with default options within SMRT Portal v2.3.0 software [1]. During the preassembly step, filtering and assembly were performed using preAssembler Filter v1 (minimum subread length, 500 bp; minimum polymerase read quality, 0.80; minimum polymerase read length, 100 bp) and preAssembler v2 (Minimum seed read length, 6,000 bp; number of seed read chunks, 6; alignment candidates per chunk, 10; total alignment candidates, 24; minimum coverage for correction, 6). The read quality was confirmed by aligning shorter reads on longer reads applying Basic Local Alignment with Successive Refinement v1 (BLASR) [5] and correcting errors using Pilon version 1.16 [6]. The chromosome and plasmid annotation performed using rapid prokaryotic genome annotation (Prokka) v1.12b [7].
The complete genome sequence of S. aureus strain JDFM SA01 consists of one circular chromosome (2,748,925 bp) with an overall GC content of 32.9% and one circular plasmid sequence (24,655 bp) with a GC content of 28.7%. A total of 2,549 predicted genes were identified on the genome, including 19 rRNA, 61 tRNA, and 2,520 coding DNA sequences (CDSs) from chromosome, while 29 CDSs were identified on the plasmid. The genomic information of S. aureus JDFM SA01 could be applied to develop new sanitation strategy for safe and high-quality dairy products.
