RESEARCH ARTICLE
Complete genome sequence of Lactobacillus amylovorus 1394N20, a potential probiotic strain, isolated from a Hanwoo calf
Young Joon Oh
1
, Joon Yong Kim
1
, Jieun Lee
2
, Seul Ki Lim
1
, Dohyeon Yu
3
, Yeon-su Oh
4
, Jinho Park
5,*
, Hak-Jong Choi
1,*
1Research and Development Division, World Institute of Kimchi, Gwangju 61755, Korea
2SME Service Department, Strategy and Planning Division, World Institute of Kimchi, Gwangju 61755, Korea
3College of Veterinary Medicine, Gyeongsang National University, Jinju 52828, Korea
4College of Veterinary Medicine and Institute of Veterinary Science, Kangwon National University, Chuncheon 24341, Korea
5Veterinary Internal Medicine, College of Veterinary Medicine, Jeonbuk National University, Iksan 54596, Korea
*Corresponding author: Jinho Park, Veterinary Internal Medicine, College of Veterinary Medicine, Jeonbuk National University, Iksan 54596, Korea., Tel: +82-63-850-0949, E-mail:
jpark@jbnu.ac.kr
*Corresponding author: Hak-Jong Choi, Research and Development Division, World Institute of Kimchi, Gwangju 61755, Korea., Tel: +82-62-610-1729, E-mail:
hjchoi@wikim.re.kr
© Copyright 2021 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Jun 22, 2021; Revised: Aug 12, 2021; Accepted: Aug 27, 2021
Published Online: Sep 30, 2021
Abstract
Lactobacillus amylovorus are known to exist in the intestinal flora of healthy cattle or pigs. The L. amylovorus strain 1394N20 was isolated from the feces of the Hanwoo calf (Bos taurus coreanae). The genome of strain 1394N20 consists of a single circular chromosome (2,176,326 bp) with overall guanine + cytosine content of 37.8 mol%. Moreover, 2,281 protein-coding sequences, 15 rRNAs, and 65 tRNAs genes were identified in the chromosome based on the results of annotation. The bacterium has a gene encoding endoglucanase, an enzyme that hydrolyzes the 1,4-β-D-glycosidic linkages in cellulose, hemicellulose, lichenin, and cereal β-D-glucans. Genomic sequencing of L. amylovorus strain 1394N20 reveals the immense potential of the strain as a probiotic with nutrient digestibility.
Keywords: Lactobacillus amylovorus; Hanwoo calf; Whole genome sequencing; Endoglucanase
According to a recent study, microbial colonization of the intestine by diverse microbiota begins before birth in mammals; however, the microbiota changes rapidly in the early postnatal life [1]. Lactobacillus, a microorganism found in the intestinal flora of mammals such as cows and pigs, has the ability to inhibit the growth of pathogenic microorganisms by lowering the colon pH, and hence is widely used as probiotics [2]. Several microorganisms use cellulose as a carbon source, which is a major component of plants. Cellulose can be hydrolyzed by cellulase, which is composed of β-1,4-glycosidic bonds, promoting the composting of organic matter and improving feed efficiency by increasing its bioavailability in the intestine of livestock [3]. The most important source of energy in ruminant diets are carbohydrates, which are major precursors of lactose and fat in milk. Microorganisms present in the rumen facilitate the use of energy from fibrinous carbohydrates, such as cellulose and hemicellulose, which are bound to fibrin and lignins present in the cell walls of plants [4].
In this study, Lactobacillus amylovorus strain 1394N20 (KCCM 12999P) was isolated from the feces of a 8-day-old healthy male Hanwoo calf. Strain 1394N20 was cultured in de Man–Rogosa–Sharpe broth (Difco, Franklin Lakes, NJ, USA) at 35°C for 24 h using the BD GasPak EZ Anaerobe Container System (Becton Dickinson Microbiology Systems, Cockeysville, MD, USA). Genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA), according the manufacturer’s instructions. The complete genome of the strain was sequenced by DNALINK (Seoul, Korea) using PacBio RSII (Pacific Biosciences, Melon Park, CA, USA). These sequences were assembled de novo using the RS Hierarchical Genome Assembly Process version 3.0 [5]. The genomes of strain 1394N20 were annotated using Pathosystems Resource Integration Center (PATRIC) version 3.6.9 [6]. Functional annotation was performed using Kyoto Encyclopedia of Genes and Genomes database (www.genome.jp/kegg) [7] and evolutionary genealogy of genes: Non-supervised Orthologous Group-mapper version 2 (http://eggnog-mapper.embl.de) [8].
The genome sequences of the strain consisted of one chromosome, with a 581.2× sequencing depth (coverage). The quality parameters for the genome assembly, namely coarse consistency (99.4%) and fine consistency (97.6%), indicated the good quality of the assembled genome evaluated using PATRIC. The complete genome of strain 1394N20 was 2,176,326 bp long, with a guanine + cytosine content of 37.8 mol%. Moreover, the chromosome comprised of 2,281 protein-coding sequences, 15 rRNA genes (five 5S, five 16S, and five 23S), 3 ncRNA genes, and 65 tRNA genes (Table 1 and Fig. 1). The genome possessed bcsZ gene encoding endoglucanase. The protein encoded by this gene is decomposed by endohydrolysis of the D-glucosidic linkage of cellulose. In addition, it contained genes including β-glucosidase (bglX), 6-phospho-β-glucosidase (bglA), and cellobiose phosphotransferase system EIIA component (celC and chbA), which converts cellobiose into D-glucose. Based on this information, it is suggested that L. amylovorus strain 1394N20 can facilitate sugar absorption by decomposing cellulose, which is difficult to digest in the intestine.
Table 1.
Genomic features of Lactobacillus amylovorus strain 1394N20
|
Properties |
Value |
|
BioProject |
PRJNA726865 |
|
BioSample |
SAMN18972587 |
|
Accession No. |
CP074196 |
|
Sequencing method |
PacBio RSII |
|
Assembly method |
HGAP v3.0 |
|
Genome size (bp) |
2,176,326 |
|
Contig |
1 |
|
Total CDSs |
2,281 |
|
rRNA genes |
15 |
|
tRNA genes |
65 |
|
G + C content (mol%) |
37.8 |
Download Excel Table
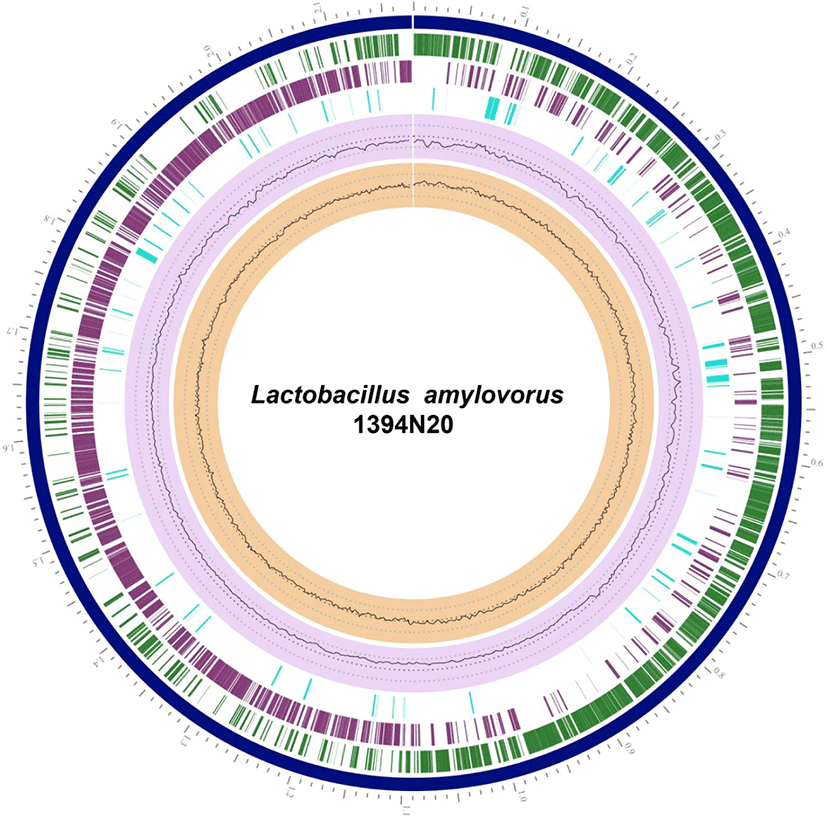
Fig. 1.
Circular view of the genome of Lactobacillus amylovorus strain 1394N20 showing the physical map of its significant features generated using PATRIC.
From outside to inside: contigs (blue), annotated reference genes (particularly, coding sequences [CDSs]) on the forward strand (green), and annotated reference genes on the reverse strand (purple). The fourth circle shows non-CDSs featured in the genome (light blue). The next circle indicates guanine–cytosine (GC) content (lavender/light purple), while the innermost circle indicates the GC skew (peach).
Download Original Figure
NUCLEOTIDE SEQUENCE ACCESSION NUMBER
The GenBank accession number for the genome of L. amylovorus strain 1394N20 is CP074196.