INTRODUCTION
Innate immunity, known as a host defense system, recognizes and eliminates pathogens. Toll-like receptors (TLRs) play a key role in detecting pathogens, such as bacteria, viruses, and chemicals as well as activating innate immunity [1,2]. TLR3 activates the inflammatory pathway by recognizing double-stranded RNA produced by viral infection. [3,4]. Inflammation maintains homeostasis and prevents further infection, but excessive inflammation also damages normal cells [5]. TLR3 activation by highly pathogenic avian influenza A H5N1 virus, pandemic H1N1 virus, and dsRNA analog polyinosinic:polycytidylic acids (Poly[I:C]) induces inflammation and impairment of lung function, and knockout of TLR3 was positive for improvement of lung innate immunity and survival rate [6,7]. Therefore, TLR3 inhibition is a good target for improving the survival rate of chickens against respiratory infectious dsRNA virus disease. Functional feed additives such as alpha-lipoic acid (ALA) inhibit TLR3 activity and improve chicken meat quality and productivity, but are not practical due to their high cost [8,9]. In chickens, it is an attractive goal to genetically understand the mechanisms of TLR3 regulation and increase its regulatory efficiency. TLR regulatory mechanisms are well established in humans, but do not apply equally in chickens. In the case of human TLR4, MD-2 protein acts as an activity inhibitor of TLR4, whereas chicken TLR4 requires complex formation with MD-2 protein for pathogen detection [10]. In the case of TLR3, the amino acid sequence of the intracellular region, Toll/interleukin-1 receptor (TIR), is markedly different in birds and primates (Fig. 1). The regulatory mechanisms of chicken TLR3 signaling remain to be studied.
In this study, we identified the pancreatic progenitor cell differentiation and proliferation factor (PPDPF) as a potential inflammatory regulator in the chicken. PPPDF is a key modulator of exocrine pancreatic development and transcriptional target of retinoic acid (RA) and pancreas transcription factor 1a (PTF1A) [11,12]. In zebrafish, PPDPF has been shown to promote pancreatic exocrine gland growth and differentiation while inhibiting pancreatic endocrine gland growth and secretion. PPDPF gene is highly overexpressed in ovarian cancers and upregulated DNA replication pathway [13]. Furthermore, the PPDPF gene was discovered in prostate tumors as a genomic marker that can be utilized to predict biochemical recurrence [14]. Liver-specific PPDPF overexpression effectively inhibits high-fat diet (HFD)-induced mechanistic target of rapamycin (mTOR) signaling activation and hepatic steatosis in mice [15]. We investigated the levels of PPDPF expression in chicken organs and TLR3-activated chicken cells, as well as the role of PPDPF in TLR3 expression and activation in chicken cells. We also explain the diversity of PPDPF across the species and suggest chicken PPDPF as a factor for TLR3 activation and inflammation.
MATERIALS AND METHODS
PPDPF protein amino acids sequences, i.e., chicken (Gallus gallus; XP_027329124.1), duck (Anas platyrhynchos; XP_027329124.1), human (Homo sapiens; Q9H3Y8.1), porcine (Sus scrofa; XP_020933363.1), bovine (Bos taurus; NP_001068762.1), equine (Equus caballus; XP_023482697.1), murine (Mus musculus; Q9CR37), and feline (Felis catus; XP_003983360) were retrieved from National Center for Biotechnology Informaion, U.S. National Library of Medicine, USA. Predicted phosphorylation site was evaluated using NetPhos-3.1 service (DTU Health Tech, Lyngby, Denmark) [16]. Predicted ubiquitination site was evaluated using RUBI Version 1.0 (BioComputing UP, Padua, Italy) [17] or obtained from neXtProt database (Swiss Institute of Bioinformatics, Lausanne, Switzerland). The structure of chicken PPDPF was predicted using IntFOLD server [18,19]. 3-dimensional (3D) structure was constructed by using PyMol software (Schrödinger, New York, NY, USA).
DF-1 chicken embryonic fibroblast cell line (Cat# CRL-12203, ATCC, Manassas, VA, USA) and the HD-11 chicken macrophage-like cell line [20] were cultured with Dulbecco’s modified Eagle’s medium (DMEM; Cat# 10-013-CVR, Corning, Corning, NY, USA) supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) in a 5% CO2 atmosphere at 37°C. The HD-11 cell line was provided by Dr. Hyun S. Lillehoj at the Agricultural Research Services (ARS) at the United States Department of Agriculture (USDA), Beltsville, Maryland, USA.
The specific-pathogen-free White Leghorn chickens (4-weeks old) were purchased from the Poultry Research Centre of the National Institute of Animal Science (NIAS; Hanoi, Vietnam). The chickens were given unlimited access to antibiotic-free feed and water. A total of 5 tissue samples were collected from the chickens, and placed in liquid nitrogen for total RNA extraction. All the experiments were conducted in compliance with the institutional rules for the care and use of laboratory animals, as well as implementing the protocol approved by the Ministry of Agriculture and Rural Development of Vietnam (TCVN 8402:2010 and TCVN 8400-26:2014).
Total RNA was isolated from the DF-1 cells, HD-11 cells, and lung tissue of White Leghorn chickens using a TRIzol reagent (Cat# 15596018, Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized using WizScript cDNA Synthesis Kit (Cat# W2202, Wizbiosolutions, Seongnam, Korea).
Real-time PCR was performed using StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with 10 ng cDNA, SYBR Green qPCR Master Mix (Cat# DQ385-40h, Biofact, Daejeon, Korea), and 1 pM primer (Table 1). Levels of TLR3, tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3), Toll-like receptor adaptor molecule 1 (TICAM1), nuclear factor-κB1 (NF-κB1), TNFα, and cyclooxygenase2 (COX2) mRNA were measured [21]. mRNA fold-change was normalized to β-actin mRNA using 2−ΔΔCt method [22].
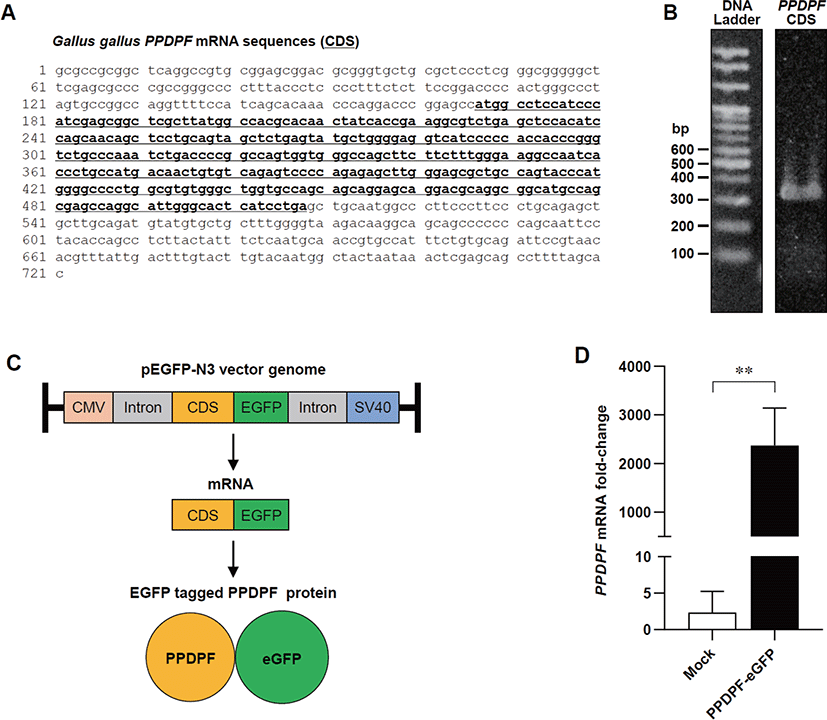
cDNA from lung tissue of White Leghorn chickens was used for PPDPF cloning. PCR was performed for isolation and cloning of PPDPF using Pfu DNA polymerase with specific primers (Table 2) under the following conditions: 95°C for 5 min, 30 cycles of 95°C 30 sec, 56°C 45 sec, 72°C 50 sec, and the final extension for 10 min at 72°C. Newly synthesized DNA fragments were purified using Gel Purification kit (Cat# K-303501, Bioneer, Daejeon, Korea). DNA fragments and pEGFP-N3 vectors were digested using HindIII and BamHI restriction enzymes. DNA fragment was inserted into the vector using the TOPcloner kit (Cat# EZ002S, Enzynomics, Daejeon, Korea) and transformed into E. coli DH5α. The plasmid was purified using the Plasmid Mini Extraction Kit (Cat# K-3030, Bioneer) and clones containing PPDPF coding sequences (CDS) were selected through sequencing (Bioneer). All processes were carried out according to the manufacturer’s instructions.
Chicken DF-1 cells were transfected with pEGFP-N3 vectors concentration at 1 µg / 1.0 × 106 cells using Lipofectamine 3000 transfection reagent (Cat# L3000008, Invitrogen) according to the manufacturer’s instructions.
Cells were washed with phosphate buffered saline (PBS; pH 7.4), fixed in 3.8% formaldehyde for 15 min, and stained with 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/mL). Fluorescence was visualized using a DMi8 fluorescence microscope (Leica, Deerfield, IL, USA) and LAS X program (Leica). Lightness and contrast of images were processed using Photoshop CC program (Adobe Systems, San Jose, CA, USA).
Cells were cultured in 96-well plates with 10% (v/v) WST-1 reagent (Cat# EZ-3000, DoGenBio, Seoul, Korea) for 2 h and measured the absorbance at 450 nm using a microplate reader (Sunrise, Tecan, Männedorf, Switzerland).
Results were obtained from three separate experiments (n = 3) and analyzed using GraphPad PRISM 8 software (GraphPad Software, San Diego, CA, USA) and Microsoft Excel software (Microsoft, Redmond, WA, USA). The data are expressed as mean ± SD. The p-value was calculated using an analysis method suitable for each experiment and specified in figure legends. Results were considered statistically significant when the p-value was < 0.05.
RESULTS
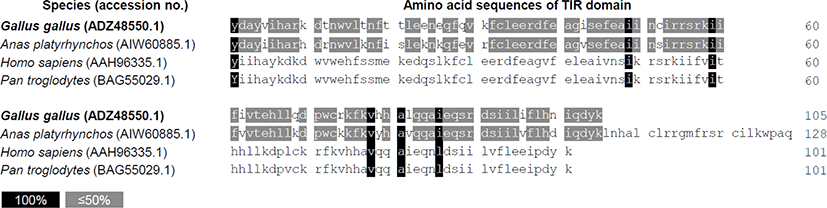
The intracellular domain TIR regulates TLR3 activity through adapter protein binding and signaling when TLR3 is activated by ligand [23]. Therefore, we identified the possibility at the amino acid sequence level for differences in the regulation of TLR3 signaling in primates and avian. Amino acid sequences of the TIR domain between two avian species, chickens (Gallus gallus, ADZ48550.1) and ducks (Anas platyrhynchos, AIW60885.1) were very similar. TIR domain sequence homology was also observed between two primates, i.e., human (Homo sapiens, AAH96335.1) and chimpanzees (Pan troglodytes, BAG55029.1). Comparison of the TIR domain sequences between avian and primates revealed very low homology (Fig. 1), suggesting the possibility that TLR3 signaling of avian species may be distinct from that of primates.
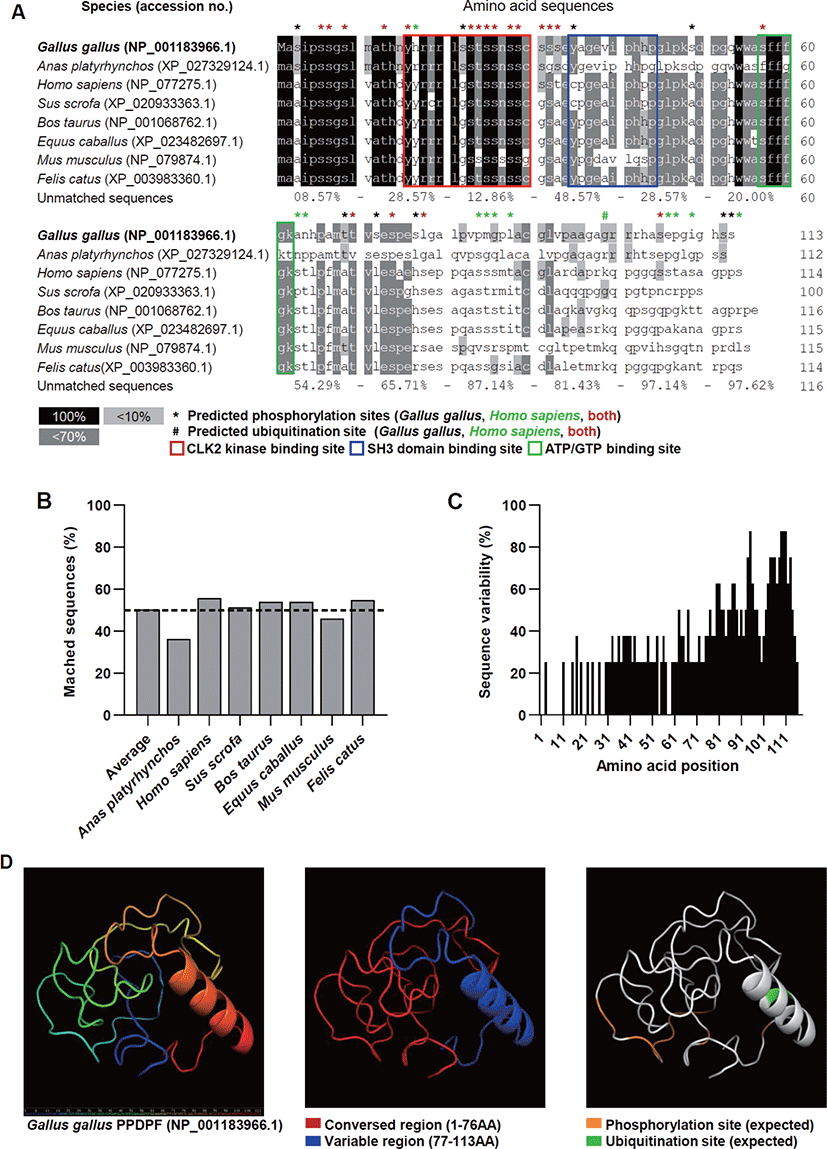
We performed an amino acid sequence-based analysis to analyze the interspecies diversity of PPDPF proteins and predict their functions. The PPDPF protein sequences were highly conserved up to the 76th amino acid (AA). The predicted ubiquitination site (99Lys) was identified only in human, bovine, equine, murine, and feline sequences (Fig. 2A). The homology of the PPDPF protein sequence to chicken PPDPF averaged only 50% in the species we investigated (Figs. 2A and B). Amino acid sequence diversity dropped sharply at 77th AA across all species investigated in this study (Figs. 2A and C). To discover the function of chicken PPDPF, the 3D structure of the chicken PPDPF protein (NP_001183966.1) was predicted using the IntFOLD server. In our protein model, chicken PPDPF was identified as a structure with one alpha-helix (Fig. 2D). In the conserved region of PPDPF (1-76AA), phosphorylation sites are located. And the CLK 2 kinase sites, a SH3 domain binding site, and adenosine triphosphate (ATP) / guanosine triphosphate (GTP) binding sites reported in PPDPF were located [11]. In the variable region (77-113AA), the alpha helix structure and the predicted ubiquitination site were located (Fig. 2D). These results suggest the possibility that PPDPF may play a role that differs from species to species.
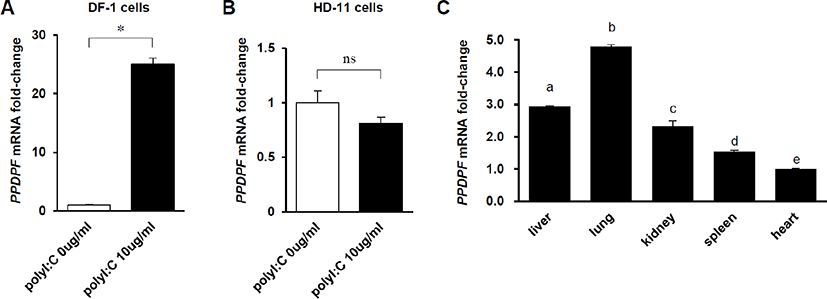
To confirm whether the expression of PPDPF in chickens is affected by dsRNA, PPDPF mRNA fold-change was measured by RT-PCR in Poly(I:C)-treated chicken cell lines. In DF-1 cells, a chicken embryonic fibroblast cell line, PPDPF mRNA expression was increased 25-fold by treatment with 10 μg/mL Poly(I:C) (Fig. 3A). And, as expected, in chicken macrophage-like cell line HD-11 cells lacking exogenous dsRNA detection ability [24], Poly(I:C) treatment did not affect the expression of PPDPF (Fig. 3B). To evaluate the PPDPF expression levels for each tissue, RT-PCR was performed with cDNA from chicken organs. The expression levels of PPDF were the highest in the lungs and the lowest in the heart among the organs which we investigated (Fig. 3C). Through these results, we confirmed the possibility that PPDPF expression was induced by Poly(I:C), a TLR3 activator, in chickens and is related to the immune response in the lungs.
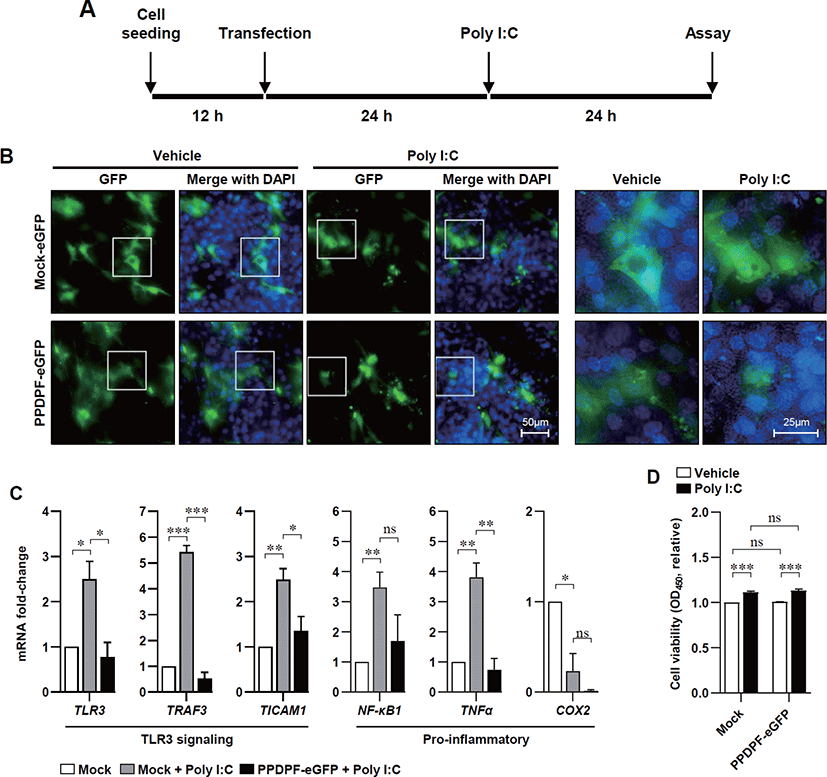
To determine the effect of PPDPF expression on TLR3 inflammatory response in chicken cells, we constructed a chicken PPDPF expression vector based on the CDS of PPDPF mRNA (accession no. NM 001197037.1) (Fig. 4A). PCR was used to isolate CDS from chicken PPDPF, which was then introduced into the pEGFP-N3 vector (Figs. 4B and C). Transfection of PPDPF inserted pEGFP-N3 vector into DF-1 cells elevated PPDPF mRNA expression, as validated by RT-PCR (Fig. 4D). DF-1 cells were cultured with Poly(I:C) for 24 hours to examine if PPDPF expression alters the expression of pro-inflammatory genes produced by Poly(I:C), and fluorescence microscopy and RT-PCR were performed (Fig. 5A). The PPDPF protein was uniformly located in the cell including the nucleus. TLR3 signaling genes (TLR3, TRAF3, and TICAM1), as well as pro-inflammatory genes (NF-B1 and TNF-α), were increased by Poly(I:C) treatment, however PPDPF overexpression suppressed TLR3, TRAF3, TICAM1, and TNF-α expression (Fig. 5C). It is of note that PPDF transfection did not induce toxicity in DF-1 cells (Fig. 5D). These results suggest that overexpression of PPDPF in chicken fibroblasts is a potential TLR3 inhibitor to downregulate the mRNA expression of TLR3-related genes induced by Poly(I:C). Through the above results, we confirmed that PPDPF inhibits Poly(I:C)-induced TLR3 mRNA expression in DF-1 cells and might act as a negative regulator for dsRNA-induced inflammation.
DISCUSSION
In chickens, TLR3 is important for immunity against major infectious diseases that threaten the avian industry, such as avian influenza virus (AIV) and Newcastle disease virus (NDV) [25,26]. But excessive inflammation might be responsible for reduced chicken productivity [27,28]. TLR-mediated inflammation against invading pathogens should be dampened to maintain homeostasis to prevent the potential damage resulted from uncontrolled responses. Numerous negative regulatory molecules have been identified and characterized at multiple levels [29]. However, no negative regulators for TLR have been reported in livestock, including poultry. In this study, we discovered PPDPF as a protein capable of suppressing TLR3-mediated inflammatory responses in DF-1 cells. The main functions of PPDPF in mammalians were well reported as a key regulator of development of pancreas in human. In contrast, the structure and function of PPDPF have not yet been reported in chickens.
In this study, we identified the potential for differences in TLR3 signaling and binding proteins in humans and chickens based on the amino acid sequence of TIR domain (Fig. 1). And we predicted the function and structure of PPDPF through bioinformatics (Fig. 2). The evolutionarily conserved domains present in chicken PPDPF proteins, the CLK2 kinase binding site, SH3 domain binding site, and ATP/GTP binding site, reveal the potential of PPDPF to participate in signaling pathways or act as enzymes. (Fig. 2A). The presence of SH3 domains in proteins suggests potential involvement in various signaling pathways by protein tyrosine kinases through protein-protein interactions [30]. CLK2 protein is implicated in a variety of signaling pathways, including regulation of inflammation and viral resistance [31,32], and cell proliferation [33]. Presence of ATP/GTP binding site in protein, it can use the energy of ATP for enzymatic action [34]. The role of the chicken PPDPF domain should be verified through a loss of function study.
We examined the relationship between TLR3 signaling and PPDPF in chicken cells. The expression level of PPDPF is upregulated by TLR3 stimulation with Poly(I:C) in DF-1 cells but not in HD-11 cells (Figs. 3A and B). HD11 cells have low reactivity to exogenous dsRNA, and TLR3 activation is completed in a very short time, so PPDPF expression may not be induced even with poly(I:C) treatment [24,35]. TLR3 is mainly located on the cell surface of non-immune cells, including fibroblasts [36]. The DF-1 chicken embryo fibroblast cell line is derived from chicken embryos and has active TLR3 signaling pathways [37,38], demonstrating that DF-1 cells might be a suitable model to study the TLR3 responses in chickens.
It is of note that, in Poly(I:C) treated DF-1 fibroblasts, PPDPF-eGFP is mainly located in the nucleus (see Fig. 5B), and mechanism behind this localization remains to be studied, but at least, it seems that PPDPF may not have direct interaction with TLR3. Mechanism behind the localization of PPDPF remains to be studied. Overexpression of PPDPF reduced pro-inflammatory gene expression in DF-1 chicken embryonic fibroblasts. These results indicate the possibility of PPDPF acting as a negative regulator of TLR3 mediated inflammation in DF-1 chicken embryonic fibroblasts. NF-κB1 can induce systemic pro-inflammatory gene expression under stress conditions, so inhibition of NF-κB1 expression by PPDPF could be effective in suppressing inflammation in poultry [39].
Future studies require evaluation of the effect of PPDPF protein on TLR3 signaling, inflammatory response, and binding protein through loss of function study of chicken PPDPF protein. Nevertheless, based on the sequence and predicted structure of the PPDPF protein and the study in PPDPF-overexpressing DF-1 cells, we suggest the possibility that PPDPF may participate in TLR3-mediated inflammatory responses in chickens.
