INTRODUCTION
Staphylococcus aureus is a commensal organism and opprtunistic pathogen frequently carried asymptomatically on the skin and mucous membranes of human and animal. Mechicillin-resistant S. aureus (MRSA) acquired resistance to methicillin and nearly all β-lactam antibiotics such as cephalosporins as a result of mecA or mecC gene acquisition [1]. Mostly, MRSA strains harbor the mecA within a large mobile genetic element known as staphylococcal casette chromosome mec (SCCmec) [2]. Besides the resistance to β-lactam antibiotics, MRSA usually exhibits multidrug resistance (MDR) phenotypes, and serious infections with such MRSA strains result in significantly enhanced morbidity and mortality [3,4].
MRSA has been known to be widespread and common, but specific genetic lineages can vary depending on geographical regions and host species [5,6]. MRSA strains adapted to humans have usually been divided into healthcare-associated (HA) and community-associated (CA) MRSA. Although the distinctions between the two groups of MRSA strains are becoming obscure, HA-MRSA and CA-MRSA have mainly been recognized in hopitalized patients and non-hospitalized population, respectively. In Korea, sequence type (ST) 5 MRSA with SCCmec type II (ST5-MRSA-II) and ST72-MRSA-IV represent the most significant clonal lineages for (HA- and CA- MRSA, respectively [7–9]. In addition to the human-adapted strains, colonization or infection with livestock-associated (LA) MRSA has been reported in several domesticated livestock animals [1,6,10]. These carrier animals not only serve as reservoirs for opportunistic infections in themselves but also can transmit LA-MRSA to other animal species or humans [10]. In this sense, the recent emergence of ST398 LA-MRSA-V and ST541 LA-MRSA-V in pigs and pig farm environments has posed a serious public health concern worldwide [11–14]. In fact, it has been reported that human workers were colonized with ST398 MRSA shortly after direct or indirect contact with pigs or carcasses in pig farms or slaughterhouses [12]. Although there is limited information on MRSA in poultry, previous studies identified two important clonal lineages of LA-MRSA strains, ST692 LA-MRSA-V and ST188 LA-MRSA-IV, in chickens, chicken carcasses, and slaughterhouse workers in Korea [15,16]. Although various types of MRSA clonal lineages, including both human- and livestock-adapted MRSA strains, have been found in humans and animals, factors affecting the transmission of human-type MRSA lineages to animals or LA-MRSA to humans are poorly understood.
One of the crucial factors in host innate immune reactions against MRSA infection or colonization is the host defense cationic antimicrobial peptide (HD-CAP) [17]. The successful transmission and adaptation of MRSA in a new host species inevitably require the ability to overcome the bactericidal action of HD-CAPs from new hosts [18–20]. Cathelicidins are small, positively charged antimicrobial peptides that constitute a unique family of HD-CAPs. Cathelicidins have been identified in neutrophils, natural killer cells, and the epithelial cells in the skin, respiratory, and gastrointestinal tracts of humans and various animals [21]. Thus, a better understanding of the potential differences in bactericidal activities of human- and animal-originated cathelicidins against human- or animal-adapted MRSA strains will facilitate new insight into host species-specific prevalence of MRSA clonotypes.
In this investigation, potential clonotype-specific resistance profiles to cathelicidins in humans (LL-37), pigs (PMAP-36), and chickens (CATH-2) were determined using six different MRSA strains derived from humans (ST5 HA-MRSA-II and ST72 CA-MRSA-IV), pigs (ST398 LA-MRSA-V and ST541 LA-MRSA-V), and chickens (ST692 LA-MRSA-V and ST188 LA-MRSA-IV). In addition, the net surface positive charges, carotenoid production, and hydrogen peroxide (H2O2) resistance profiles of the MRSA strains were analyzed to identify phenotypic determinants linked to cathelicidin resistance.
MATERIALS AND METHODS
A total of 45 MRSA isolates of human and animal origin were used in this investigation (Table 1). All human isolates were selected from two previous studies from our laboratory: eight ST5 HA-MRSA and eleven ST72 CA-MRSA isolates [22,23]. Nine ST398 LA-MRSA and six ST541 LA-MRSA isolates were selected from a previous investigation of LA-MRSA in healthy pigs [11]. Six ST692 LA-MRSA and six ST188 LA-MRSA isolates from healthy broiler chickens were selected and obtained from the national surveillance of antimicrobial resistance in livestock performed during 2018-2019 by the Korean Centers for Disease Control and Prevention. This study was conducted under protocols approved by the Institutional Animal Care and the Use Committee of Chunag-Ang University, Anseong, Korea (Approval No. 2018-00112).
MRSA, methicillin-resistant Staphylococcus aureus; MLST, multilocus sequence typing; SCCmec, staphylococcal cassette chromosome mec; OX, oxacillin; MIC, minimum inhibitory concentration; ST, sequence type; HA, halthcare-associated; AMP, ampicillin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; FOX, cefoxitin; PEN, penicillin; FUS, fusidic acid; GEN, gentamicin; RIF, rifampicin; CA, community-associated; CHL, chloramphenicol; TET, tetracycline; SYN, quinupristin-dalfopristin; SXT, sulfamethoxazole-trimethoprim.
For isolation of MRSA, swab samples were inoculated into 4 mL of tryptic soy broth (TSB; Difco Laboratories, Detroit, MI, USA) containing 10% NaCl and incubated 18–20 h at 37°C with shaking at 200 rpm. Next, 20 μL of the enriched cultures were streaked onto chromID MRSA SMART agar (bioMérieux, Marcy-l’Étoile, France), then grown for 18–20 h at 37°C. Suspected MRSA colonies from each sample were selected and subcultured on tryptic soy agar (Difco Laboratories) for further identification. To confirm if they were indeed S. aureus, all 45 isolates were subjected to 16S ribosomal RNA sequencing [24] and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF; Microflex, Daltonics Bruker, Bremen, Germany). Briefly, presumptive S. aureus colonies were applied onto the matrix and the samples were then ionized in an autmated mode with a laser beam in the MALDI-Biotyper Realtime Classification system. The peptide mass fingerprints of bacterial samples were used to identify S. aureus (score values of ≥ 2.0) based on the spectral database (MALDI Biotyper 3.1). All identified MRSA isolates were then grown in Mueller-Hinton broth (Difco Laboratories) or TSB (Difco Laboratories), depending on each experiment.
The antimicrobial susceptibility of MRSA isolates was examined using the standard disc diffusion assay according to the 2018 Clinical and Laboratory Standards Institute (CLSI) guidelines [25]. The 13 antimicrobial drugs used in the disc diffusion assays were ampicillin (AMP, 10 μg), cefoxitin (FOX, 30 μg), penicillin (PEN, 10 μg), gentamicin (GEN, 10 μg), clindamycin (CLI, 2 μg), chloramphenicol (CHL, 30 μg), erythromycin (ERY, 15 μg), mupirocin (MUP, 200 μg), sulfamethoxazole-trimethoprim (SXT, 23.75/1.25 μg), rifampicin (RIF, 5 μg), quinupristin-dalfopristin (SYN, 15 μg), tetracycline (TET, 30 μg), and ciprofloxacin (CIP, 5 μg). Mupirocin discs were obtained from Oxoid (Hampshire, UK), and the remaining antimicrobial discs were purchased from BD BBLTM (Becton Dickinson, Franklin Lakes, NJ, USA). A standard E-test® (bioMérieux) method was used to determined the minimum inhibitory concentrations (MICs) of oxacillin (OXA) according to the manufacturer’s protocol. The MIC values of MRSA isolates were determined according to the CLSI M100 and VET08 documents [26,27]. Two reference strains, S. aureus MW2 and S. aureus ATCC® 29213, were included in the antimicrobial susceptibility assays.
All confirmed MRSA strains were subjected to multilocus sequence typing (MLST) as described before [28]. MLST has widely been accepted for a moleuclar epidemiological method of sequence-based typing in S. aureus, which analyzes seven relatively conserved houskeeping genes that encode essential proteins. The seven target loci (aroE, arcC, glpF, tpi, gmk, pta, and yqiL) were amplified via polymerase chain reaction (PCR) and sequenced. The STs were then determined as suggested in the MLST database (http://pubmlst.org/saureus/) [28]. The allelic profiles of each STs of MRSA strains were: ST5 (1-4-1-4-12-1-10), ST72 (1-4-1-8-4-4-3), ST398 (3-35-19-2-20-26-39), ST541 (3-35-19-60-20-26-39), ST692 (12-89-1-1-4-5-90), and ST188 (3-1-1-8-1-1-1). The types of SCCmec were determined through a series of multiplex PCR analyses, as previously described [2]. SCCmec types were assigned based on combinations of ccr (ccrA1-3, ccrB1-4, and ccrC) and mec gene complexes [2,24]. For spa typing, the spa repeat regions were PCR-amplified and sequenced to examine the tandem repeats, and spa types were determined in each MRSA isolate based on the SpaServer database (http://spa.ridom.de/) [29]. The agr types (I-IV) of MRSA isolates were assinged through a PCR-based method, as described previously [29].
Three different HD-CAPs of LL-37, PMAP-36, and CATH-2 origin were used to assess the genotype- and host-specific susceptibility patterns of MRSA isolates to LL-37 [30], PMAP-36 (porcine myeloid antimicrobial peptide) [31], and CATH-2, respectively [32]. Human cathelicidin LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES) was obtained from Peptide International (Louisville, KY, USA). PMAP-36 (GRFRRLRK KTRKRLKKIGKVLKWIPPIVGSIPLGCG) and CATH-2 (RFGRFLRKIRRFR PKVT ITIQGSARF) were synthesized at GL Biochem, Shanghai, China with a purity of >90%. Based on the manufacturer’ analytical data such as reverse-phase high performance liquid chromatography (HPLC) or MALDI-TOF mass spectrometry (MS), lyophilized peptides were resuspended in phosphate-buffered saline (PBS), aliquoted, and stored at −20°C.
In vitro susceptibility assays to LL-37, PAMP-36, and CATH-2 were performed as previously described using RPMI medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% Luria-Bertani (LB) broth [33]. Briefly, overnight cultures of S. aureus cells grown in Tris buffered saline (TBS) were collected, washed two times with RPMI media, and then adjusted to ~1 × 104 CFU/mL in the RPMI containing 10% LB broth. Next, a final S. aureus inoculum of ~5 × 103 CFUs was incubated at 37°C in the presence of LL-37 (10 mg/mL), PMAP-36 (1.0 mg/mL), or CATH-2 (0.5 mg/mL). After 3 h incubation with peptides, aliquots (0.1 mL) of samples were processed for quantification of surviving bacterial cells (CFU/mL) by plating 10-fold serial dilutions of each culture on TSA plates. The peptide concentrations were adopted from extensive preliminary assays showing their inability to completely eliminate the viability of the initial bacterial inocula over the 3-h experiment period. Data are shown as the mean % of surviving MRSA CFUs ± SDs of cathelicidin-exposed versus unexposed bacterial cells. Three independent experiments were performed in triplicates for each MRSA isolate.
To assess the relative staphylococcal cell surface positive charge that can affect susceptibility to HD-CAPs, cytochrome c binding assays were carried out on the 45 MRSA strains as described before [34,35]. Briefly, MRSA isolates were grown overnight (16–18 h) in TSB, washed twice with 20 mM morpholinepropanesulfonic acid (MOPS, pH 7.0) buffer, and resuspended in MOPS at an optical density (OD)600nm of 1.0. Staphylococcal cells were then incubated with 50 μg/mL of cytochrome c (Sigma-Aldrich) for 15 min at room temperature. Next, the reaction mixture was centrifuged (10,000×g) for 1 min to pellet the bacterial cells, and the quantity of free cytochrome c that remained in the supernatant was determined by measuring the OD530nm. A higher concentration of unbound cytochrome c in the supernatant corresponds to a more positively charged staphylococcal cell surface [36]. Three independent experiments were performed for each MRSA isolate.
MRSA isolates were cultured in TSB to the stationary growth phase (24 h) at 37°C with shaking at 200 rpm to quantify carotenoid production. After incubation, staphylococcal cells were pelleted, washed three times with PBS, and then diluted in PBS to ~1.0×109 CFU/mL. The carotenoid contents of the MRSA isolates were collected using the methanol extraction method as previously described [37], and then quantified spectrophotometrically by measuring the OD462nm. At least three independent assays for carotenoid quantification were performed for all MRSA isolates.
Susceptibility to hydrogen peroxide was determined as previously described by Liu et al. [38]. Briefly, ~2.0×109 CFUs of MRSA isolates were incubated with H2O2 (1.5% final concentration) at 37°C for 2 h, and catalase (1,000 U/mL, Sigma-Aldrich) was added to remove residual H2O2. Ten-fold dilutions were then prepared and placed on TSA plates for the enumeration of surviving staphylococcal cells. Data are expressed as the mean % of surviving CFU ± SDs of H2O2-treated versus untreated cells.
The quantitative data obtained in this study were analyzed for statistical significance using the Kruska-Wallis analysis of variance (ANOVA) test with the Tukey post hoc correction for multiple comparisons. Statistical significance of experimental data was set at p values < 0.05.
RESULTS
A total of 45 MRSA isolates derived from humans (n = 19), pigs (n = 14), and chickens (n = 12) were used in this study (Table 1). All human and pig isolates were selected from previous studies [11,22,23]: eight isolates of ST5 HA-MRSA-II, eleven isolates of ST72 CA-MRSA-IV, eight isolates of ST398 LA-MRSA-V, and six isolates of ST541 LA-MRSA-V. As shown in Table 1, MLST analyses revealed that 12 chicken-associated MRSA isolates were ST692 LA-MRSA-V and six ST188 LA-MRSA-IV. Except for eight ST5 HA-MRSA-II isolates, which belonged to agr type II, all other 37 MRSA isolates were agr type I.
As presented in Table 1, three (t002, t2460, and t601) and five (t324, t13921, t664, t2461, and t148) different types of spa were identified in ST5 HA-MRSA-II and ST72 CA-MRSA-IV isolates, respectively. Unlike the various spa types observed in CA-MRSA and HA-MRSA isolates, all animal isolates tended to have ST-type-specific spa sequences. Except for one ST398 LA-MRSA (t571), the other ST398 LA-MRSA isolates belonged to t18102 or t18103. All six ST541 LA-MRSA isolates belonged to spa type t034. The chicken-derived ST692 and ST188 LA-MRSA isolate groups belonged to t2247 and t189, respectively (Table 1).
In general, the antimicrobial resistance patterns of MRSA strains differed depending on their ST and spa type. All LA-MRSA isolates showed MDR phenotypes to three or more subclasses of antimicrobial agents (Table 1). In particular, three isolates of ST398 LA-MRSA-V with spa type t18103 displayed the highest level of MDR (> 8 subclasses of antibiotic agents). In contrast to human-associated MRSA isolates (ST5 and ST72 MRSA isolates), all MRSA isolates of livestock origin (ST398, ST541, ST692, and ST188 MRSA isolates) were resistant to tetracycline. Interestingly, three ST types (ST398, ST692, and ST188) of LA-MRSA strains were resistant to ciprofloxacin, whereas all six ST541 LA-MRSA strains were susceptible to ciprofloxacin. In agreement with previous reports [39], ST5 HA-MRSA strains displayed higher levels of MDR than ST72 CA-MRSA isolates.
When grouped into six different STs (ST5, ST72, ST398, ST541, ST692, and ST188) of strain groups, the ST5 HA-MRSA strains had the highest oxacillin MIC values (512 mg/mL), followed by ST692 LA-MRSA (32–256 mg/mL), ST188 LA-MRSA (128 mg/mL), ST72 CA-MRSA (32–64 mg/mL), ST541 LA-MRSA (16–32 mg/mL), and ST398 LA-MRSA (4–24 mg/mL) (Table 1).
To assess the potential genotype-specific differences in susceptibility to HD-CAPs of human and animal origins among the six ST groups of MRSA isolates, in vitro 2-h survival assays were carried out for all 45 MRSA isolates against LL-37 (10 μg/mL), PMAP-36 (1.0 μg/mL), or CATH-2 (0.5 μg/mL). Interestingly, the ST5 HA-MRSA, ST692 LA-MRSA, and ST188 LA-MRSA isolates exhibited overall higher survival profiles against LL-37 than the ST72 CA-MRSA, ST398 LA-MRSA, and ST541 LA-MRSA isolates (Fig. 1A). The two STs of chicken origin, ST692 and ST188 MRSA isolates, displayed similar survival levels against LL-37. However, human- and pig-associated MRSA isolates exhibited significantly different levels of resistance to LL-37 between the two host-specific ST types (ST5 MRSA > ST72 MRSA, p < 0.01; ST398 > ST541 LA-MRSA isolates, p < 0.05).
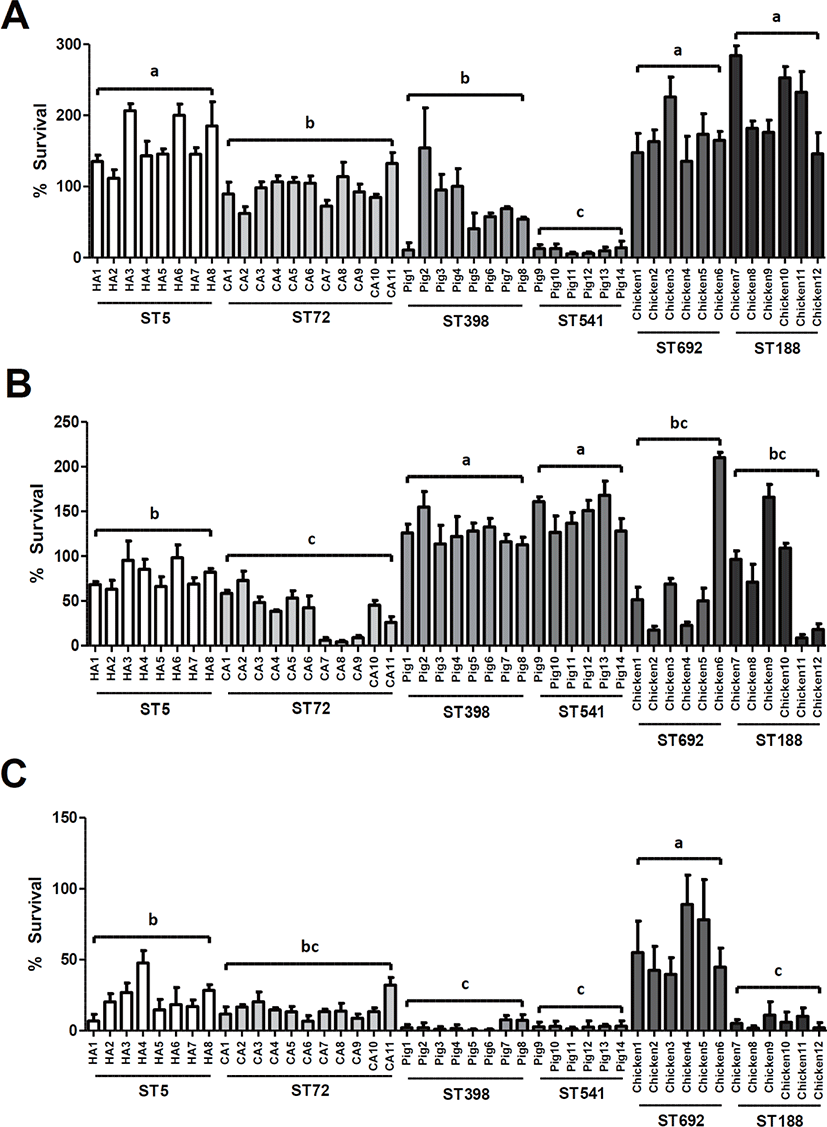
Susceptibility assays to PMAP-36 revealed that pig-associated LA-MRSA isolates were significantly more resistant to porcine cathelicidin than the human-associated or chicken-associated MRSA isolates (p < 0.01) (Fig. 1B). Although there were no significant differences in susceptibility to PMAP-36 between the two STs of LA-MRSA isolates (ST398 versus ST541 or ST692 versus ST188), ST5 HA-MRSA isolates exhibited significantly higher survival than ST72 CA-MRSA isolates (p < 0.01).
As shown in Fig. 1C, similar to the host-specific resistance to PMAP-36 observed in ST398 LA-MRSA strains isolated from pigs, ST692 LA-MRSA strains derived from chickens showed the highest level of resistance to the CATH-2 among the six ST groups of MRSA isolates. In contrast, ST188 LA-MRSA displayed significantly lower levels of resistance to CATH-2 than the ST692 LA-MRSA strains (p < 0.01). The two groups of swine-associated MRSA strains, ST398 LA-MRSA and ST541 LA-MRSA, showed the lowest levels of resistance to the CATH-2 peptide compared to the human- and chicken-associated MRSA strains (Fig. 1C).
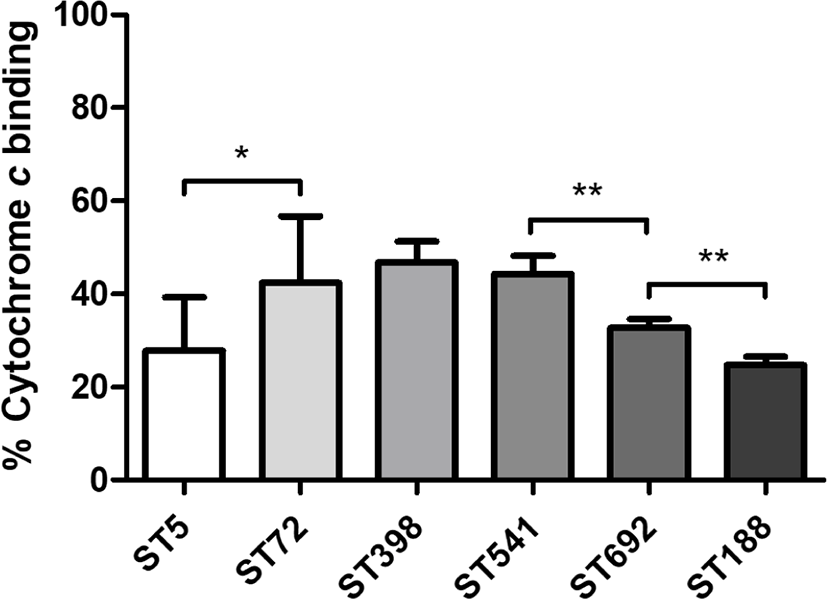
Cytochrome c binding assays revealed that ST5 HA-MRSA and ST188 LA-MRSA had the highest net surface positive charge levels among the six ST groups of MRSA strains (Fig. 2). Unlike the human- and chicken-associated MRSA isolates, which exhibited significant differences between ST5 HA-MRSA and ST72 CA-MRSA (p < 0.05) or ST692 vs. ST188 LA-MRSA (p < 0.01), swine-associated LA-MRSA isolates did not show significant differences in cell surface charge between the ST398 and ST541 MRSA groups.
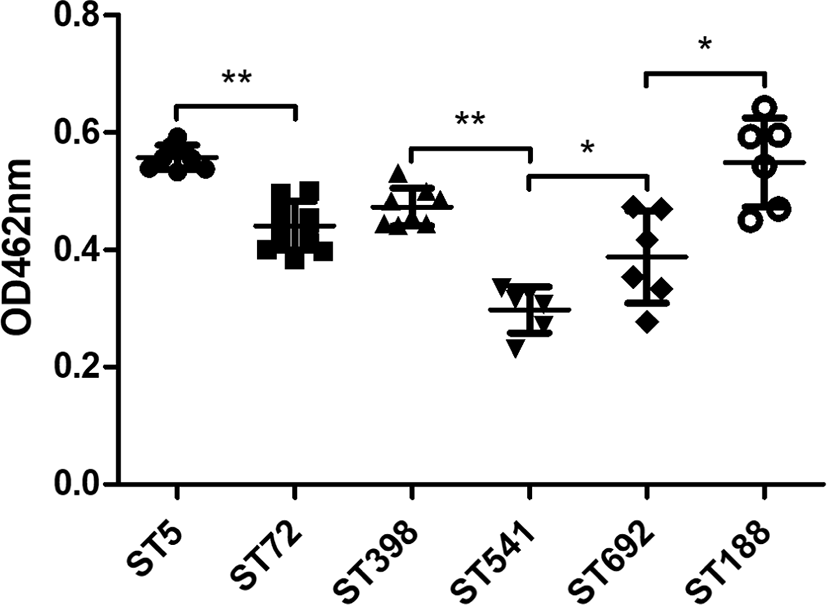
Quantification of carotenoid production in all 45 MRSA strains revealed that ST5 HA-MRSA and ST188 LA-MRSA strains exhibited the highest carotenoid content among the six different ST groups (Fig. 3). When the strain groups were compared within their host species, ST5 HA-MRSA, ST398 LA-MRSA, and ST188 LA-MRSA showed significantly higher levels of carotenoid production than the ST72 CA-MRSA, ST541 LA-MRSA, and ST692 LA-MRSA strains (p < 0.05).
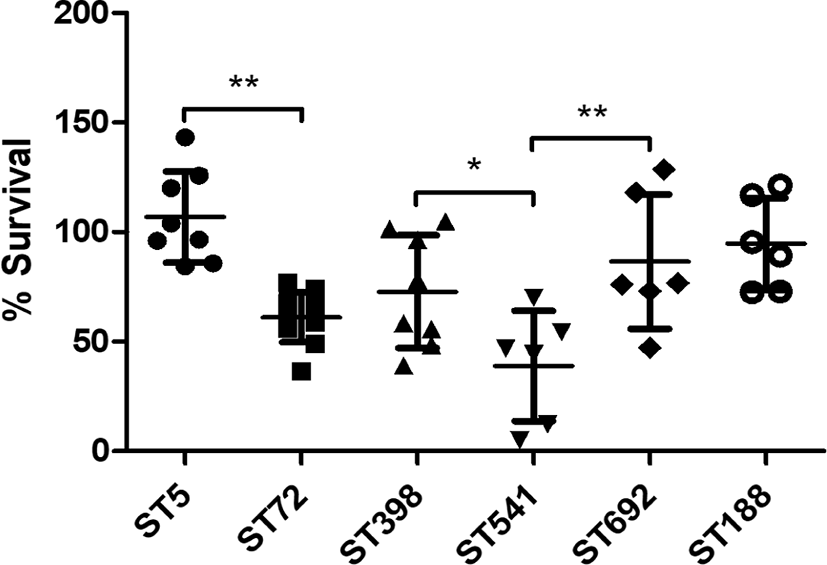
As shown in Fig. 4, in vitro H2O2 susceptibility assays over a 2-h period revealed that the ST5 HA-MRSA and ST398 LA-MRSA strain groups exhibited significantly higher survival in the presence of 1.5% H2O2 than that of ST72 HA-MRSA and ST541 LA-MRSA strains (p < 0.05). In contrast, there was no significant difference in the mean survival against H2O2 between the two chicken-derived MRSA strains, ST188 and ST692. Of note, ST5 HA-MRSA strains showed the highest level of resistance to H2O2 among the six different ST groups.
DISCUSSION
MRSA is an important human and animal pathogen that can cause frequent and often life-threatening infections in both community and hospital settings [4,40]. However, the mechanisms involved in the transmission of human- or animal-adapted MRSA strains to new host species remain poorly understood. One of the critical components in host defense against MRSA infections involves innate immune responses, including HD-CAPs such as cathelicidins [41–43]. Since MRSA infections in human and animal hosts are caused mainly by colonizing strains of staphylococci [3,44], the pathogen needs to deploy survival and adaptation mechanisms against the cathelicidins of human and animal hosts.
It has been shown that certain clonal lineages of MRSA strains are found in limited numbers of animal species or geographical regions [1,6,10]. Some recent studies have reported that ST72-MRSA-IV and ST5-MRSA-II represent major community and hospital MRSA clones in Korea, respectively [7–9]. In addition to the human-adapted MRSA clones, several major LA-MRSA clones have been recognized in livestock animals and meat production chains in Korea. The nationwide spread and prevalence of ST398-MRSA-V and ST541-MRSA-V in pigs and pork production systems in Korea have been demonstrated in several previous studies [11,13,14,16]. Furthermore, ST692 and ST188 LA-MRSA strains have also been isolated from chicken and chicken meat production chains in Korea [15,16,21]. The two groups of chicken-derived LA-MRSA clones used in this study were ST692-MRSA-V with spa type t2247 and ST188-MRSA-IV with spa type t189 (Table 1).
Both host and bacterial factors may play critical roles in the adaptation and transmission of MRSA within or between human and animal hosts [19,40,45]. In this regard, one of the crucial innate immune responses of human and animal hosts against newly colonizing MRSA strains is HD-CAPs, especially cathelicidins. A previous report from our laboratory revealed that ST5 HA-MRSA-II tended to show higher resistance to LL-37 and bovine myeloid antimicrobial peptide-28 (BMAP-28), suggesting a potential role of cathelicidin resistance in enhancing the virulence and mortality of ST5 HA-MRSA-II [22]. In line with these observations, ST5 HA-MRSA-II displayed significantly higher levels of resistance against LL-37 and PMAP-36 origins than the ST72 CA-MRSA-IV strains (Figs. 1A and 1B). The two chicken-derived LA-MRSA clones, ST692 and ST188 LA-MRSA, displayed higher levels of resistance to LL-37 andthan the ST72, ST398, and ST541 MRSA strains (p < 0.01), while ST398 and ST541 exhibited the highest levels of resistance to the porcine cathelicidin PMAP-36 (p < 0.01). Besides being prevalent in poultry farms, ST188 MRSA strains have been recognized as major CA-MRSA in Australis and China [46,47]. ST692 MRSA has also been identified in slaughterhouse workers in Korea [16], indicating that the higher levels of resistance to LL-37 in ST188 and ST692 MRSA may help to colonize human host. The observation that ST692 MRSA isolates are significantly more resistant to CATH-2 than ST188 MRSA isolates is likely to be correlated with the higher prevalence of the ST692 in chicken farms in Korea [15]. ST398 MRSA has been isolated from pigs and pig farm environment in many countries including Korea [5,7,10,11,13]. Of note, the clonal lineage of ST541 MRSA has been detected in healthy pigs in Korea [11]. As shown in Fig. 1B, ST398 and ST541 MRSA isolates were more resistant to the cathelicidin of pig origin (PAMP-36), correlating with previous reports of predominant colonization of pigs with CC398 (ST398 and ST541) strains. However, significantly enhanced LL-37 resistance in ST398 might be associated with higher prevalence of ST398 versus ST541 MRSA strains [11]. Overall, these data indicated that MRSA strains of human or animal origin tended to show enhanced levels of resistance to the cathelicidins of the same host species from which they were derived.
Several recently published reports have demonstrated that the HD-CAP resistance phenotype in staphylococci is associated with physicochemical alterations in the cell membrane, such as increased net cell surface positive charge, altered cell membrane fluidity, and fatty acid composition [48–51]. Consistent with previously published data, the ST5 HA-MRSA-II strains exhibited an enhanced net surface positive charge than the ST72 CA-MRSA-IV strains [22], indicating that the increased net surface positive charge in the ST5 HA-MRSA-II strains contributed to the enhanced resistance to cathelicidins (Fig. 2). However, although the pig-derived LA-MRSA strains exhibited the highest level of resistance to PMAP-36 (Fig. 1B), these two ST groups (ST398 and ST541) had lower levels of net surface positive charge than the human (ST5)- and chicken-derived (ST692 and ST188) MRSA strains (Fig. 2). The ST188 MRSA strains also showed significantly elevated surface positive charges compared to the ST692 MRSA strains (p < 0.01), correlating with the LL-37 resistance phenotype (Fig. 1A), but not with CATH-2 resistance (Fig. 1C). The enhanced surface positive charge in S. aureus has been associated with altered expression/sequence variation of dltABCD, mprF, and graRS genes [34,35,48]. In addition, degree of O-acetylation in peptidoglycan cell wall has been proposed to be involved in antimicrobial peptide resistance via surface positive charge regulation in S. aureus [48]. These additional factors affecting surface positive charge in S.aureus need to be addressed in future studies. Moreover, aside from the surface positive charge, cell envelope perturbations such as celll membrane fluidity, altered cell membrane permeabilization by cathelicidins, and staphylococcal memebrane fatty acid composition need to be evaluated. Based on the results, it is likely that there are multiple mechanisms involved in the host- and clonotype-specific cathelicidin resistance profiles of MRSA strains in addition to the net cell surface positive charge, especially for LA-MRSA strains of chicken and pig origins.
Carotenoid pigmentation in S. aureus has been shown to affect cell membrane fluidity and counteract oxidative host defense mechanisms. It has been demonstrated that increased carotenoid biosynthesis could contribute to HD-CAP resistance in S. aureus by decreasing cell membrane fluidity [51]. In agreement with previous reports [22], ST5 HA-MRSA strains with enhanced resistance to the three cathelicidins (Figs. 1A, 1B, and 1C) exhibited significantly increased levels of carotenoid production compared to the ST72 CA-MRSA strains (Fig. 3). In addition, correlating with the LL-37 resistance profiles (Fig. 1A), the ST5 HA-MRSA and ST188 LA-MRSA strains produced significantly higher carotenoid contents than the other four groups of MRSA strains (p < 0.05). Enhanced carotenoid production in S. aureus has previously been linked to enhanced resistance to human antimicrobial peptides and cell wall targeting antimicrobial agents [18]. However, although the pig-associated LA-MRSA strains (ST398 and ST541) exhibited the highest levels of resistance to PMAP-36 (Fig. 1B), these strain groups did not produce higher levels of carotenoids than the ST5, ST72, and ST188 strain groups (Fig. 3), indicating that changes in cell membrane fluidity via carotenoid production have a very limited impact on the PMAP-36 resistance phenotype in ST398 and ST541 strains. Furthermore, the significant difference in carotenoid production observed between ST692 and ST188 LA-MRSA strain groups did not correlate with a higher level of CATH-2 resistance in the ST692 strain group versus the ST188 strain group (Fig. 1C). Collectively, these results suggest that human-type MRSA and LA-MRSA strains deploy different strategies to counteract the bactericidal activity of HD-CAPs encountered during host adaptation and colonization.
Given the impact of carotenoid production on the antioxidant activity of MRSA against the host innate immune response [38,52,53], in vitro susceptibility to H2O2 was determined for the six clonotypes of MRSA strains. Consistent with the carotenoid pigmentation profiles (Fig. 3), the ST5 HA-MRSA and ST398 LA-MRSA strain groups exhibited significantly higher levels of resistance to H2O2 than the ST72 CA-MRSA and ST541 LA-MRSA strains, respectively (Fig. 4). Although the results failed to reach statistical significance in resistance to H2O2, higher levels of carotenoid production in ST188 MRSA strains were also correlated with the moderate increase in H2O2 resistance compared to the ST692 strains. In a previous study it has been suggested that the carriage of superantigen genes in ST5 HA-MRSA may play a role in the higher virulence versus ST72 CA-MRSA [54]. Based on our result it is likely that enhanced resistance to HD-CAPs and H2O2 via increased surface positive charge and carotenoid production also contribute to the higher virulence of ST5 HA-MRSA versus ST72 CA-MRSA strains. Recently, ST188 S. aureus has been identified as a major clonal lineage causing infections in multiple host species in China [47]. Whole genome sequence analyses revealed that ST188 S. aureus strains can be transmitted more easily among different host species [47]. The higer level of LL-37 resistance (Fig. 1A), enhanced surface positive charge (Fig. 2) and higher carotenoid production in ST188 MRSA strains (Fig. 3) versus ST692 MRSA strains may help to colonize diverse host species. On the contrary, the highest level of CATH-2 resistance observed in ST692 MRSA strains may be linked to the prevalence of this lineage in chickens [15].
In conclusion, the results of the present investigation suggest that: (i) there are differences in the biological activities of human- and animal-derived cathelicidins against MRSA strains of human (ST72 CA-MRSA and ST5 HA-MRSA), pig (ST398 and ST541 LA-MRSA), and chicken (ST692 and ST188 LA-MRSA) origins; (ii) ST5 HA-MRSA strains have higher resistance to various types of cathelicidins than ST72 CA-MRSA strains, as correlated with an increased net surface positive charge, enhanced production of carotenoid pigments, and enhanced resistance to H2O2; (iii) in contrast to human-type MRSA strains, pig- and chicken-associated MRSA strains had cathelicidin resistance mechanisms other than surface positive charge regulation and carotenoid production; and (iv) clonotype-specific cathelicidin resistance profiles may contribute to increased virulence or adaptation within new host species. To the best of our knowledge, this study is the first to evaluate potential role of HD-CAP resistance in human and animal host adaptation of MRSA strains.
