INTRODUCTION
Economically, it is important that different breeds of livestock can be easily identified. Consumers often encounter processed products, including meats, at markets and it is necessary to identify the origin, breed, and species of the animals used in products. Several Korean studies have described tools for determining the breed of Korean native chicken (KNC; Gallus gallus domesticus) used in various products [1,2]. However, the current traceability system in Korea only considers chicken meat and egg quality. The ability to discriminate between different chicken breeds using a genetic approach could improve consumer confidence while also safeguarding unique genetic resources.
Yeonsan Ogye, one of the KNC breed, is characterized by black feathers, skin, and bones, and considered an important element of Korean heritage. Globally, only a few chicken breeds display similar black plumage to Yeonsan Ogye, including Ayam Cemani from Indonesia, H’mong from Vietnam, and Svarthöna from Sweden [3,4]. In general, the techniques used for identifying specific chicken breeds are based on morphological characteristics, but it is sometimes challenging to morphologically distinguish breeds with similar phenotypes.
Genetic information could be applied for precise breed identification. Various genetic markers have been developed and used to obtain genetic information. Typically, microsatellite (MS) markers are utilized for the identification of various livestock breeds [5–7]. However, as MS markers have unique characteristics, they are not always reflective of the entire genome, and some also have high mutation rates [8]. In addition, research using MS markers requires significant human input, and the interpretation of the results is subjective.
Single nucleotide polymorphism (SNP) markers could overcome the limitations of MS markers [9]. Recently, genotyping methods using SNP arrays have been developed over several generations, and the cost of genotyping continues to fall. Hence, a large amount of SNP data is available for application as genotype biomarkers and can rapidly provide accurate information for breed identification. However, identifying optimal SNP markers for specific populations using high-density SNP chips is still quite complex.
Machine learning using classification models is possible to deal with the large genotype data effective. The classification model is a process of distinguishing the class of new input data based on learned data with labels through various algorithms. In particular, the Random Forest (RF) and AdaBoost (AB) algorithms are effectively used to reduce overfitting, handle large data, and select the important variables.
The objective of this study was to determine optimal SNP marker combinations to discriminate a target chicken population (Yeonsan Ogye) from other breeds using two machine learning algorithms (RF and AB).
MATERIALS AND METHODS
This research has been approved by the Institutional Animal Care and Use Committee (IACUC) of Chungnam National University (202103A-CNU-061).
An overview of the procedure used for identifying SNP markers to discriminate the Yeonsan Ogye breed is provided in Fig. 1.
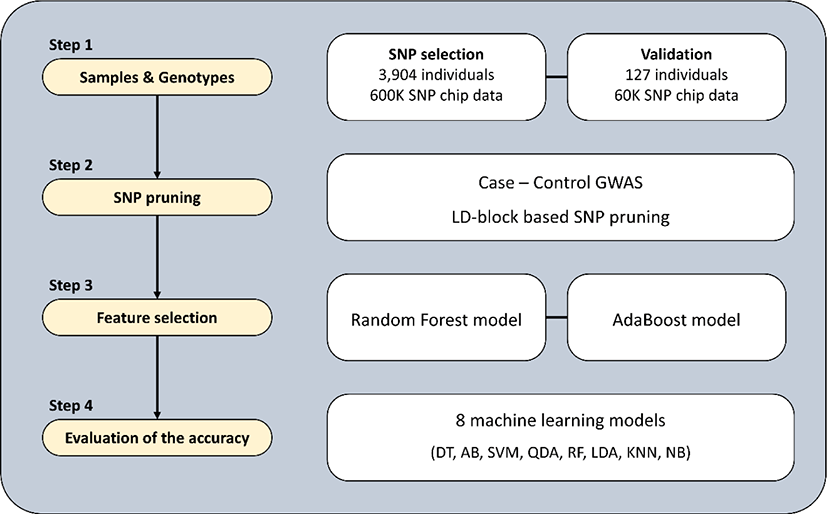
Three data sets were used in this study: Sets 1 and 2 for selecting SNP markers, and Set 3 for validation (Table 1). Sets 1 and 2 consisted of 3,904 individuals from 198 chicken breeds, genotyped with a 600K SNP array (Affymetrix, Santa Clara, CA, USA) [10]. Set 1 constituted populations of KNC from the Korean National Institute of Animal Science (NIAS), including Yeonsan Ogye (189 birds), and other indigenous (208 birds from five lines) and adapted KNC (218 birds) breeds. Set 2 consisted of commercial chickens (CC; 34 broilers and 20 layers) and various other global chicken breeds from the SYNBREED project in Germany [11]. The SYNBREED dataset included 3,235 individuals and 174 breeds from 32 countries, including Africa, South America, Asia, and Europe. Set 3 consisted of Yeonsan Ogye (67 birds) and KNC (30 birds from two lines), genotyped using a custom 60K SNP array made by our research team, and an F2 generation crossbreed population of Yeonsan Ogye and White Leghorn (30 birds) genotyped with an Illumina 60K SNP array (Illumina, San Diego, CA, USA) [12].
A total of 542,717 common SNPs was derived from Sets 1 and 2, and there were two major quality control (QC) cut-offs: genotyping rate ≥ 90% and minor allele frequency ≥ 0.05. For determining Yeonsan Ogye-specific SNPs, the derived SNPs were subjected to a case-control genome-wide association study (GWAS) performed using PLINK 1.9 software [13]. In that analysis, the case group was the Yeonsan Ogye population, and the control group comprised all other populations. The significant SNPs were figured out based on the Bonferroni-corrected p-value (α = 0.01). The linkage disequilibrium (LD) was calculated, and LD block-based SNP pruning was conducted to select one SNP per 50 LD blocks.
Machine learning was applied for the feature selection of pruned SNP markers to reduce the number of SNP markers and identify optimal markers. Feature importance values were calculated through two machine learning models: RF using the “randomForest” package in R software [14] and AB using the “adabag” R package [15]. SNPs with importance values higher than the point at which feature importance rapidly decreased were classified as optimum markers. Principal component analysis (PCA) was conducted to verify the SNP marker selections.
To resolve data imbalances before analysis, only one individual was randomly selected from each of the 197 populations in the control group. To confirm the accuracy of discrimination for the Yeonsan Ogye chicken population, 70% of the total data were used as the training set, and the remaining 30% as the test set, based on five repeated 10-fold cross-validation. Eight different machine learning models were employed to evaluate the accuracy: Decision Tree (DT), AB, Support Vector Machine (SVM), Quadratic Discriminant Analysis (QDA), RF, Linear Discriminant Analysis (LDA), K-Nearest Neighbor (KNN), and Naïve Bayes (NB) [16–18]. Principle components 1 (RF, 47.4%; AB, 45.3%) and 2 (RF, 5.9%; AB, 5.4%) values, derived from the PCA for marker selection, were used to build these eight classification models with the “caret” R package [19].
For performance verification, each machine learning model was assessed based on confusion matrix values: accuracy, specificity, sensitivity (recall), precision, and F1-score.
Where TP is true-positive (number of correct predictions for the case group), TN is true-negative (number of correct predictions for the control group), FP is false-positive (number of incorrect predictions for the case group) and FN is false-negative (number of incorrect predictions for the control group).
Validation tests were conducted on independent populations to validate the discriminatory performance of the selected marker combinations. Set 3 was used for validation analysis; the data were genotyped using 60K SNP arrays. Minimac3 and Minimac4 software were used for data imputation prior to the analysis [20].
RESULTS
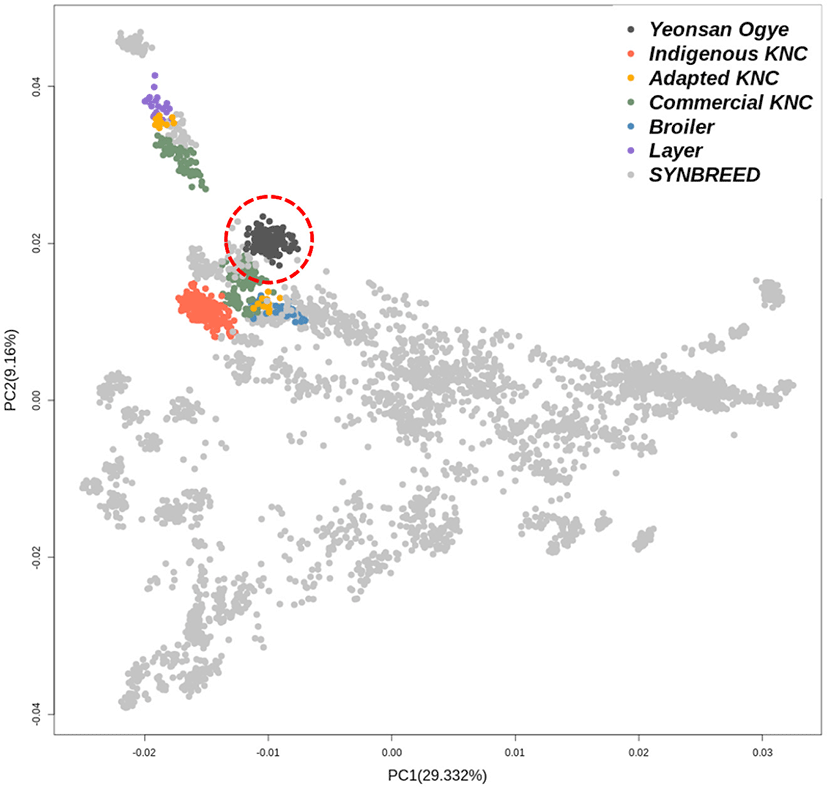
PCA of the 600K SNP genotype data for the entire population was performed. Fig. 2 shows the genetic clustering for each population. The indigenous KNC populations were clustered separately from the other groups, while the adapted KNC populations tended to cluster with CC such as broilers and layers. Contrary to this, the Yeonsan Ogye population was well differentiated from both the SYNBREED and Korean populations.
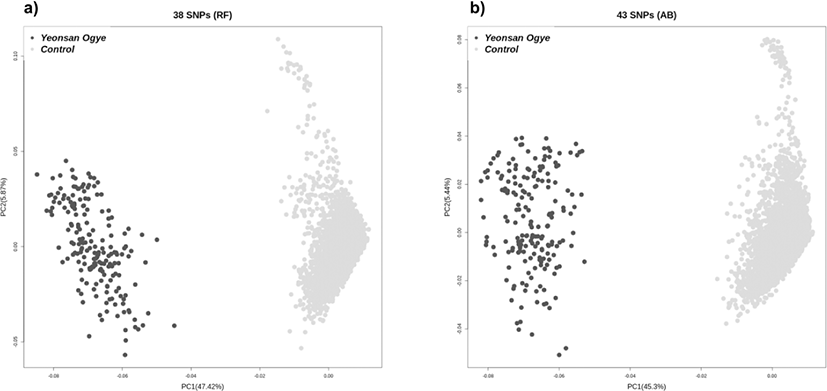
A case-control GWAS was performed to determine significant SNP markers. The target breed, Yeonsan Ogye, was the case group, and the other populations comprised the control group. The GWAS revealed 285,227 significant SNPs based on a Bonferroni corrected p-value of < 0.01. As well as LD blocks, 100,799 haplotype blocks were distinguished. Ultimately, 120 SNPs were extracted through LD-based SNP pruning of 151,062 markers common to both the GWAS results and LD blocks. In a final step, 38 (RF) and 43 (AB) SNPs were identified as the optimal marker combinations. According to the PCA of these SNP combinations, the Yeonsan Ogye population was accurately distinguished from the control group species (Fig. 3).
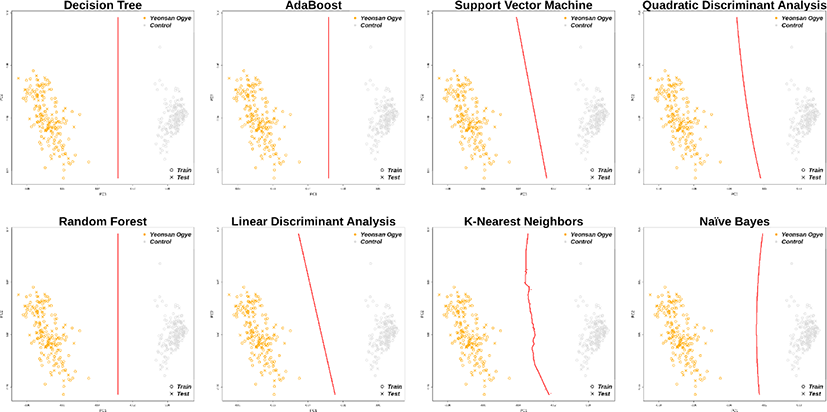
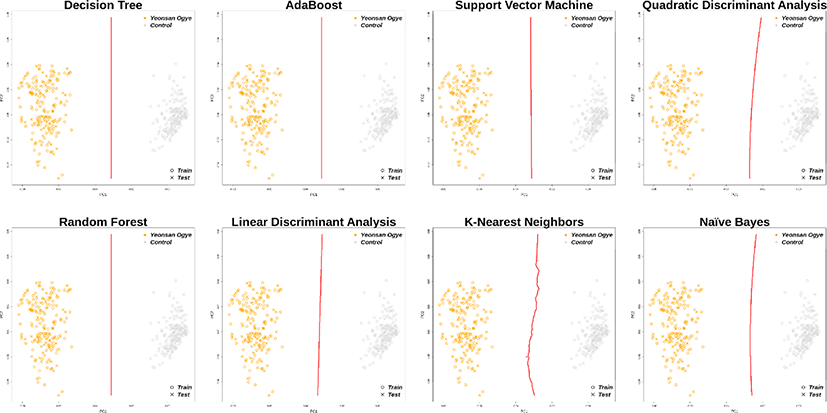
Using the 38 and 43 optimal SNP combinations described above, all eight machine learning algorithms discriminated the Yeonsan Ogye population perfectly (Fig. 4 and 5) according to the confusion matrix values (i.e., accuracy = 1.00) (Table 2).
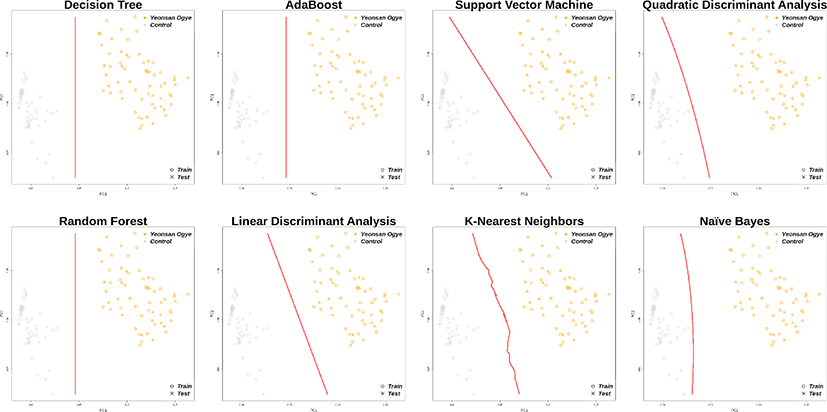
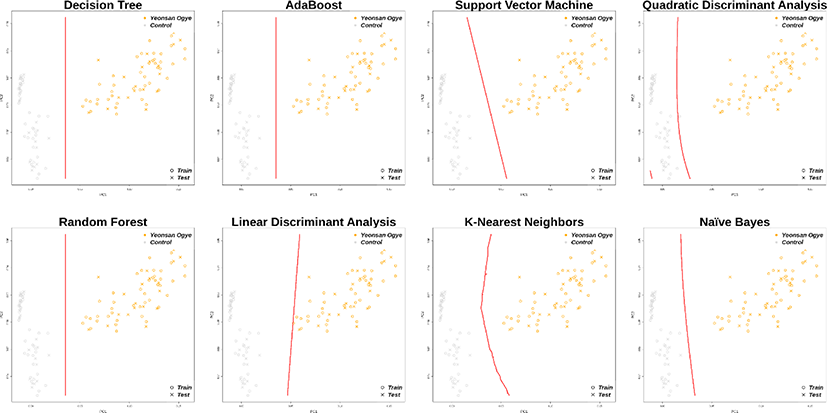
In total, 30 markers from the imputation results overlapped with the previously selected marker combinations, and distinguished the Yeonsan Ogye and control group populations accurately; the confusion matrix values were all 1.00 (Fig. 6 and 7), except for that of QDA (0.97) based on AB feature selection.
DISCUSSION
Optimal strategies for breed identification are essential for protecting livestock pedigree, and for industrial research. Native chickens are a particularly important target for biodiversity conservation; chickens are able to adapt well to new environments [21]. Park et al. [22] reported that the provision of breed information for native chickens promoted consumption.
Genotyping methods have been developed over several generations, and the cost of genotyping continues to decline. Hence, extensive genotype data are available for use as biomarkers. SNP markers have been used for genetic classification based on PCA, F-statistics, and genotype frequencies [23–25]. However, identifying optimal SNP markers to identify specific breeds using high-density SNP chips is still quite challenging.
In this study, several markers were identified based on GWAS and LD pruning results and using high-density 600K SNP chip data. Johnson et al. [26] and Wallace et al. [27] explained that it is challenging to determine whether genetic markers identified through GWAS are causative genes in response to LD. Bakshi et al. [28] stated that more informative results can be obtained by removing SNPs with strong LD relationships from the analysis. In our analysis, the target breed, Yeonsan Ogye, was effectively discriminated using SNP markers selected with consideration of LD.
Machine learning is an artificial intelligence technology for classifying data and making predictions. We applied machine learning algorithms to identify SNP marker combinations for Yeonsan Ogye classification through GWAS and LD pruning. Machine learning has been used to select SNP markers for various livestock species [29–32]. Moreover, applying feature selection to GWAS results can reduce dimensionality and overfitting errors when identifying markers, resulting in more accurate predictions [33].
In this study, RF and AB models were used to determine optimal SNP marker combinations; 38 and 43 significant SNP markers were identified, respectively, and both sets showed remarkable classification power. Notably, 14 SNPs were shared between the two marker sets, and it was possible to differentiate the target population with sufficient accuracy (more than 98%) using those markers. In addition to accuracy, other confusion matrix evaluation indices, such as sensitivity (recall) and precision, also demonstrated the high classification power of the marker combinations.
The precise results obtained herein could be explained by the fact that the Yeonsan Ogye chicken is a genetically unique breed. The PCA plot of the 600K genotype data showed that the Yeonsan Ogye population was clustered separately from the other breeds. Further, Yeonsan Ogye chicken had a gene pool independently from the entirely black chickens in the SYNBREED group, such as Cemani and Sumatran from Indonesia, and Silkies from China.
The marker combinations identified for the Yeonsan Ogye pure line (PL) showed impressive results in the validation test. Two of five KNC lines and the Yeonsan Ogye-White Leghorn crossbreed were included in the control group for the validation test. The 30 SNPs were common to both SNP marker sets and correctly differentiated KNC and Yeonsan Ogye, as also seen during the SNP marker selection process. The Yeonsan Ogye and White Leghorn crossbreeds were also clearly distinguished; the phenotypes of the individuals comprising this F2 generation were very diverse. The marker combinations showed the ability to perfectly discriminate pure Yeonsan Ogye birds, even from other chicken breeds with a similar phenotype.
Generally, the chickens available on the market are CC produced by using PLs through three-or four-way crossbreeding. Since breed-specific markers are identified using PLs, the applicability to breeds that have not been verified via the marker selection process is limited. Although verification analysis was performed on the crossbreeds in this study, it would be complicated to apply it to crossbreeds other than White Leghorn. Ultimately, the discriminatory power of the optimal SNP marker combinations identified herein must be verified through application to other populations.
