INTRODUCTION
The taxonomical composition of the bacterial community in the colon of a healthy adult pig is Firmicutes 35%, Bacteroidetes 21%, Proteobacteria 3%, and Spirochetes 2%. In the jejunum and ileum, 70% of the bacteria are Proteobacteria and 20% are Firmicutes. On the other hand, more than 75% are Firmicutes and about 13% are Proteobacteria in the cecum and colon. The administration of a complex lactobacilli cocktail (Lactobacillus johnsonii and Lactobacillus mucosae, isolated from healthy pig feces) reduces the number of potential entero-pathogens such as Clostridia and E. coli promoting a healthy gut in pigs [1,2].
Berry fruits such as strawberries, raspberries, and blackberries contain very high levels of polyphenols, particularly ellagitannins (cloudberry 315.1 mg/100 g of fw, red raspberry 297.3 mg/100 g of fw, strawberry 77.1 mg/100 of fw, sea buckthorn 1 mg/100 g of fw). Although ellagitannins and ellagic acid have very low bioavailability, they are metabolized as urolithins (3,4-benzocoumarin derivatives) by gut microbiota, and are circulated in plasma in the form of glucuronide and sulfate conjugates. Urolithins have anti-inflammatory, antioxidant, antimicrobial, anticarcinogenic, and antiglycative effects. Berry-derived polyphenols increase beneficial gut bacteria such as Akkermansia muciniphila and decrease pathogenic gut microbiota. In the clinical studies by Bouyahya et al. [3], blueberry consumption increased Bifidobacterium spp. (B. spp.) and Lactobacillus acidophilus (L. acidophilus) and decreased Bacteroides and Clostridium spp. (C. spp.).
One of the primary functions of the intestinal immune system is to maintain immune homeostasis between the nonpathogenic gut microbiome, harmless exogenous food antigens, pathogenic microbes, and damage-assigned self-antigens. T cells can be differentiated into CD4+ T cells and CD8+ T cells. Naive CD4+ T cells can be differentiated into Th1, Th2, Th9, Th17, Th22, Treg cells and follicular helper T (Tfh) cells. T helper cells induce and control immune responses as part of adaptive immunity. Th1 cells induce phagocytosis and intracellular killing of microbes, mainly by releasing proinflammatory cytokine interferon (IFN)-γ, while Th2 cells induce specific responses to extracellular pathogens and cell-mediated inflammation, mainly by releasing anti-inflammatory cytokine interleukin (IL)-10 and IL-4. Th1 and Th2 cells are regulated mutually, and Th1/Th2 balance is important for removing infections. T-box expressed in T cells (T-bet) and GATA binding protein 3 (GATA3) are Th1 and Th2 polarizing transcription factors, where T-bet suppresses GATA3 transcription and function through direct protein-protein interaction, and GATA3 suppresses signal transducer and activator of transcription 4 (STAT4) expression [4].
IL-17-releasing cells, Th17 cells, show high expression of retinoid acid-related orphan receptor γt (RORγt) which is a transcriptional regulator. Cytokine IL-17 plays an essential role in neutrophil and macrophage recruitment and mediates the inflammatory response to infectious agents. Forkhead box protein P3 (FOXP3+) regulatory T cells are a specialized population of CD4+ T cells, which are essential for limiting immune activation and preventing systemic autoimmunity. Treg cells from the thymus derive the tolerogenic response to microbiota and food antigens in the intestine. This process occurs in gut training lymph nodes through interacting with antigen-presenting cells that convert circulating naïve T cells into Treg cells. Dysregulation of Treg cells causes many chronic inflammatory disorders, including inflammatory bowel disorder [5].
Our group investigated the effects of R. coreanus by-products on the gut microbiota and immune system of rats to confirm their efficacy as an animal feed additive [6]. We found that immune modulation effect by inducing T cell polarization was enhanced in the mesenteric lymph nodes (MLNs) from the rats fed with RC byproducts. In this study, fermented feed was prepared using R. coreanus-derived lactic acid bacteria (L. plantarum GBL 16, 17). The purpose of this study was to investigate the effect of combination with lactobacillus fermented feed and conventional probiotics (Bacillus subtills, Aspergillus oryzae and Yeast) on the composition of intestinal microbes and the regulation of intestinal immune homeostasis in pigs.
MATERIALS AND METHODS
All experimental protocols involving animals in the present study were approved by the Institutional Animal Care and Use Committee (IACUC) of Gyeongnam National University of Science and Technology (No. 2018-5).
The R. coreanus byproducts used in this experiment were made with the method of Yu et al. [6] who suggested that the remaining peel and seeds after making R. coreanus extracts be dried in an oven at 50°C for 24 hours and pounded into a fine powder. L. plantarum GBL 16 and 17 separated from R. coreanus were subcultured more than two times in Man, Rogosa and Sharpe (MRS) medium (Difco, Detroit, MI, USA) to ferment the R. coreanus byproducts [7]. In order to make R. coreanus-derived LAB fermented feed (RC-LAB fermented feed), R. coreanus-derived LAB (L. plantarum GBL 16 and 17, 109 CFU/g) liquid culture was added to equal 20% of a medium composed of R. coreanus byproducts 20%, molasses 13%, corn 15%, and soybean meal 32%, and then solid fermented (25 ± 3°C for 4 to 5 days), dried (less than 12% moisture content), and powdered to a size of 60 mesh. RC-LAB fermented feed (L. plantarum 106 CFU/g) and Bacillus subtills (108 CFU/g, EUNJIN BIO, Cheonan, Korea) 0.2%, Aspergillus Oryzae (107 CFU/g, EUNJIN BIO) 0.2%, Yeast (108 CFU/g, EUNJIN BIO) 0.2% was mixed to make a fermented probiotic feed.
Data were collected from July to 2018 to September 2018 in the Dasan Genetics farm (Namwon, Korea), which has the temperature-controlled close facilities through a ventilation system. Artificial light was provided from 8:00 to 18:00. Seventy-two finishing Berkshire pigs were randomly allotted (at approximately 65 kg body weight) to four different treatment groups (A: barrows fed with the basal diet; B: barrows fed with 0.3% RC-LAB fermented feed + probiotics; C: gilts fed with the basal diet; and D: gilts fed with 0.3% RC-LAB fermented feed + probiotics). The animals were fed ad libitum the assigned diet and water and tested for 75 days with eighteen replicates per treatment group. Body weights were measured twice at the beginning and end of the experiment. The dietary treatment was either a commercial finishing pig diet based on corn and soybean meal (control groups; A and C) or the diet supplemented with 0.3% RC-LAB fermented feed + probiotics (treatment groups; B and D). The corn-soybean meal-based commercial pig diet contained 3,306 kcal of metabolizable energy/kg, 0.88% standardized ileal digestibility of lysine, 0.4% total calcium, and 0.28% digestible phosphorus. Both dietary treatments were formulated to meet or exceed the requirements of the National Research Council [8] of finishing pigs and to have comparable metabolizable energy, crude protein (CP), standardized ileal digestibility of lysine, total calcium, and digestible phosphorus levels (Table 1).
Provided the following quantities of minerals per kg of complete diet: Se, 0.1 mg; I, 0.3 mg; Mn, 24.8 mg; Cu·SO4, 54.1 mg; Fe, 127.3 mg; Zn, 84.7 mg; Co, 0.3 mg.
Ileal contents were collected for each group after harvest to extract the genomic DNA of the intestinal microorganisms in the pigs. Genomic DNA from the ileal content samples was extracted using Fecal DNA MiniPrepKitsTM (Zymo Research, Irvine, CA, USA) and the sequence was analyzed via next-generation sequencing using the Illumina Miseq platform (Illumina, San Diego, CA) and CLcommunityTM software [9].
qPCR was then used to analyze the gut microbes containing gut short-chain fatty acids (SCFAs)-producing bacteria with the extracted DNA using a Rotor-Gene SYBR® Green PCR Kit (Qiagen, Hilden, Germany) and Rotor-Gene Q (Qiagen) [6]. Primers of 12 types of intestinal microbes are shown in Table 2. Pigs were sacrificed to analyze the intestinal immune homeostasis regulation effects of the RC-LAB fermented feed with probiotics. MLNs and spleen were separated from the pigs, and a TRIzol® (Life Technologies, Paisley, UK) reagent was added to them. Then it was homogenized with SilentCrusher M (Heiodlph, Germany). RNA was separated and cDNA was synthesized according to the manufacturer’s instructions using a PrimeScript™ One Step RT-PCR Kit Ver.2 (TAKARA BIO, Otsu, Tokyo). RT-PCR was performed using a GeneAMP® PCR System 9700 (Thermo Fisher Scientific, Waltham, MA, USA). The mRNA expression of transcription factors and cytokines was used to analyze T-cell polarization. The separation and analysis of RNA from the spleens and MLNs and the cycling conditions for all primers, qPCR, and RT-PCR were done according to the methods reported by Yu et al. [6]. Relative mRNA expression was normalized to glycolytic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression levels. T1, T2, T3 and Treg-cells transcription factors and cytokines primers are shown in Table 3.
Adapted from Da Yoon Yu et al. with CC-BY-NC [6].
Adapted from Da Yoon Yu et al. with CC-BY-NC [6].
RESULTS AND DISCUSSION
The effect of feeding 0.3% RC-LAB fermented feed with probiotics on body weight gain is shown in Table 4. The results show that there was no effect on treatment group body weight gain, especially between groups A and B, and between C and D. Bunte et al. [10] and Min et al. [11] reported that two operational taxonomic units (OTUs) belonging to genus Lactobacillus and Bifidobacterium in the feces and digesta of the small and large intestines significantly increased, but did not affect body weight gain when liquid feed fermented with L. plantarum, Pediococcus pentosaceus, and Lactococcus lactis was fed to pigs.
The effect of RC-LAB fermented feed and probiotics on the intestinal beneficial microbiota composition in pigs was investigated (Table 5). The intestinal microbiota composition of the pigs consisted of Firmicutes 88.58%, Proteobacteria 5.42%, Bacteroidetes 3.77%, and Actinobacteria 1.62% at the phylum level. In humans and rats, Firmicute and Bacteroidetes are dominant bacteria [12], but Firmicute and Proteobacteria were identified as prevalent bacteria in this study. Looft et al. [13] have reported that Bacteroidetes, Firmicutes, and Proteobacteria dominated porcine fecal bacterial communities at the phyla level. However, they found that Proteobacteria increased and Bacteroidetes decreased when performance-enhancing antibiotics (chlortetracycline, sulfamethazine, and penicillin [known as ASP250]) were fed to pigs. Obese mice have fewer Bacteroidetes and more Firmicutes in their feces compared to lean mice. This shift of the fecal bacterial community is related to the higher energy-harvesting capacity of the obese mice. Also, a higher growth rate in pigs is related to a higher feed conversion rate [13].
A, barrows fed with the basal diet; B, barrows fed with 0.3% RC-LAB fermented feed; C, gilts fed with the basal diet; D, gilts fed with 0.3% RC-LAB fermented feed.
The relative abundance of phyla Firmicutes, which was the most dominant phyla in the pigs’ intestines of this study, was not different among the treatment groups (A, B, C, D), but that of class Bacilli was 12.05% higher in the treatment groups (B, D; 24.55% on average) than in the control groups (A, C; 12.5% on average) (p < 0.05). Genera Lactobacillus, Streptococcus, Weissella, Pediococcus, Leuconostoc, which belong to class Bacilli, were all significantly higher in the treatment groups (B, D) than in the control groups (A, C) (p < 0.05). In particular, genera Lactobacillus and Streptococcus increased by 8.51% and 4.68% on average in the treatment groups B and D, respectively. The relative abundance of class Clostridia, which accounts for most of phyla Firmicutes, significantly decreased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05), but genera Howardella significantly increased in the control group (A) within the barrow groups (A, C) (p < 0.05). The relative abundance of class Negativicutes was not significantly different between the control groups (A, C) and the treatment groups (B, D), but genera Mitsuokella was significantly higher in the barrow treatment group (B) compared to the barrow control group (A) (p < 0.05).
Important probiotic microorganisms in genus Lactobacillus include LAB species (L. acidophilus, Lacticaseibacillus casei, L. gasseri, Limosilactobacillus reuteri, and L. helveticus) and Enterococcus faecalis, Enterococcus faecium, Lactococcus lactis, Leuconostoc mesenteroides, Pediococcus acidilactici, Sporolactobacillus inulinus, and Streptococcus thermophilus species. Probiotics have the ability to maintain a proper balance between pathogens and beneficial bacteria to prevent gastrointestinal infections and disorders, and also have an immunomodulatory action on the host, which helps remove cholesterol.
Lactic acid (LA), one of the LAB metabolites, impairs the pH gradient between cytosol (alkaline) and the external environment (acidic), and dissipates membrane potential to achieve antimicrobial activity through the destruction of pathogenic cells. Bacteriocins are heat- and acid-resisting LAB oligopeptides, which have antimicrobial activity against pathogenic bacteria and fungi [14]. Clostridium clusters XIVa and IV are predominant intestinal bacteria, accounting for 10% to 40% of the total bacteria, and are well known as indispensable regulators of internal homeostasis [15].
As weaned pigs age, the relative abundances of 7 bacterial genera (Fibrobacter, Collinsella, Roseburia, Prevotella, Dorea, Howardella, and Blautia), including Howardella, significantly increase. The functional maturation of this gut bacterial community is associated with the digestive system, glycan biosynthesis and metabolism, and vitamin B biosynthesis [16]. When comparing low-birthweight (LBW) piglets to normal-birth-weight piglets, LBW piglets have lower rates of Faecalibacterium at three days of age, Flavonifractor at seven days of age, Lactobacillus, Streptococcus, and Prevotella at twenty-one days of age, and Howardella at twenty-one and thirty-five days of age. Metabolomics analysis suggests that these lower rates are related to fatty acid metabolism, amino acid metabolism, as well as bile acid biosynthesis [17]. L. reuteri ZLR003 affects the fecal microbiota composition of piglets, which is associated with the metabolism of SCFAs, long chain fatty acids, and free amino acids (FAAs). In particular, genera Mitsuokella and Megasphaera have significant positive effects on serum FAAs content [18].
The relative abundance of phyla Proteobacteria, the second most dominant phyla in porcine intestines, was not different between the control and the treatment groups (A, B, C, D), but classes Betaproteobacteria, genera Parasutterella and Sutterella were significantly higher in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05). Genera Parasutterella sp. (Parasutterella excrementihominis species, family Sutterellaceae, order Burkholderiales, class Betaproteobacteria, and phylum Proteobacteria) is a core component of human and animal gut microbiota and known to be asaccharolytic, a succinate producer, and a high L-cysteine consumer. In response to the carbohydrate-rich diet, genera Parasutterella sp. activates the fatty acid biosynthesis pathway, resulting in increased body weight gain and obesity development [19]. Dietary fat intake (total fatty acids, saturated fatty acids, trans fatty acids, monounsaturated fatty acids, polyunsaturated fatty acids, n3-FAs, and n6-FAs) affects the bacterial composition and structure, and Sutterella, Fusobacterium, and Tyzzerell increase with fatty acid intake.
Phyla Bacteroidetes and classes Bacteroidia, which is the third dominant group of the porcine intestines in this study, showed no difference between the control and the treatment groups (A, B, C, D). However, genera Prevotella, Bacteroides, Prevotellaceae_uc significantly increased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05). The Prevotella abundance in the pig gastrointestinal tract (GIT) varies depending on the growth stage. It is less abundant in the suckling and nursery stages and is more dominant in the growing and finishing stages after weaning. After post-weaning, Prevotella-enriched gut microbiota can improve the growth performance of pigs because it increases energy harvested by fermenting complex dietary polysaccharides [20].
For phyla Actinobacteria and classes Actinobacteria_c, there was no difference between the control and the treatment groups (A, B, C, D). However, genera Bifidobacterium significantly decreased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05). Actinobacteria are gram-positive bacteria with high guanine and cytosine (GC) DNA content, and they belong to one of the largest bacteriological phyla. Actinobacteria have an extensive secondary metabolism and produce about two-thirds of all naturally derived antibiotics used in clinics [21]. Bifidobacteria and Lactobacilli are dominant members of the gut microbiota of suckling pigs and are a probiotic bacterial species with various immunomodulatory properties [22].
The effect of RC-LAB fermented feed with probiotics on the intestinal microbiota composition was analyzed for harmful microorganisms. There was no difference between the control and the treatment groups (A, B, C, D) in the relative abundance of phyla Firmicutes (Table 6). There also was no difference between barrows (A) and gilts (C) control groups in the relative abundance of class Clostridia, which accounts for most of the phyla Firmicutes. However, the relative abundance of class Clostridia in the controls (A, C) and the treatment groups (B, D) was on average 66.34% and 39.29%, respectively, meaning there was a significant decrease of 27.05% in the treatment groups (B, D) (p < 0.05). Clostridium, Terrisporobacter, Romboutsia, and Asaccharospora at a genus level significantly decreased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05). In particular, the relative abundance of Clostridium, Terrisporobacter, and Romboutsia decreased by 14.41%, 16.88%, and 6.31% on average in the treatment groups (B, D). Genus Terrisporobacter was significantly lower in the gilts control group (C) compared to the barrows control group (A), which means there is a significant difference between sexes (p < 0.05). Clostridium spp. has about 200 distinct species, in which Clostridium is a heterogeneous group with pathogenic and probiotic characteristics. Terrisporobacter is an emerging anaerobic pathogen, and Romboutsia and Asaccharospora are closely related to genus Clostridium.
A, barrows fed with the basal diet; B, barrows fed with 0.3% RC-LAB fermented feed; C, gilts fed with the basal diet; D, gilts fed with 0.3% RC-LAB fermented feed.
The relative abundance of class Erysipelotrichi was significantly higher in the gilts treatment group (D) compared to the gilts control group (C) (p < 0.05). However, genus Kandleria was significantly lower in the gilts treatment group (D) compared to the gilts control group (C) (p < 0.05), and genus Sharpea was significantly lower in both the barrows and the gilts treatment groups (B, D) (p < 0.05). Although the relative abundance of class Negativicutes was no different between the control and the treatment groups, genus Megasphaera and Dialiste were significantly lower in the treatment groups (B, D) (p < 0.05).
The relative abundance of phyla Proteobacteria was not different between the control and the treatment groups, but class Epsilonproteobacteria was significantly lower in both treatment groups (B, D) (p < 0.05). Genus Campylobacter was also significantly reduced in the treatment groups (B, D) (p < 0.05). Genus Helicobacter was significantly reduced in the barrows treatment group (B) (p < 0.05). The relative abundance of class Gammaproteobacteria and genus Escherichia was significantly lower in both treatment groups (B, D) (p < 0.05). The relative abundance of genus Campylobacter, Helicobacter, and Escherichia in the phyla Proteobacteria was significantly lower in both treatment groups (B, D) (p < 0.05). In particular, Escherichia showed that the averages in the control groups (A, C) and the treatment groups (B, D) were 3.84% and 0.99%, respectively, which was a decrease of 2.85% on average in the treatment groups (B, D).
Presti et al. [23] confirmed Erysipelotrichi as a potential biomarker for irritable bowel syndrome (IBS), and Gammaproteobacteria, Enterococcus, and Enterococcaceae as potential biomarkers for inflammatory bowel diseases (IBD). Family Erysipelotrichaceae is abundant in the intestinal tract of mammals and is associated with host metabolic disorders, inflammatory diseases, and the concentration of N-acetylgalactosamine. Further research will be needed on the functional roles of Erysipelotrichaceae related to hosting phenotypes in the future [24]. The relative abundance of phyla Actinobacteria and class Actinobacteria_c had no difference between the control and the treatment groups, but genus Corynebacterium significantly decreased in the treatment groups (B, D) (p < 0.05). Corynebacterium is a diverse genus with industrial, medical or biological importance. Corynebacterial species are commensals, but some are notable pathogens containing the human pathogen Corynebacterium diphtheriae and the animal pathogen Corynebacterium pseudotuberculosis. Corynebacterium ulcerans is a zoonotic pathogen, which people often acquire from canine pets [25].
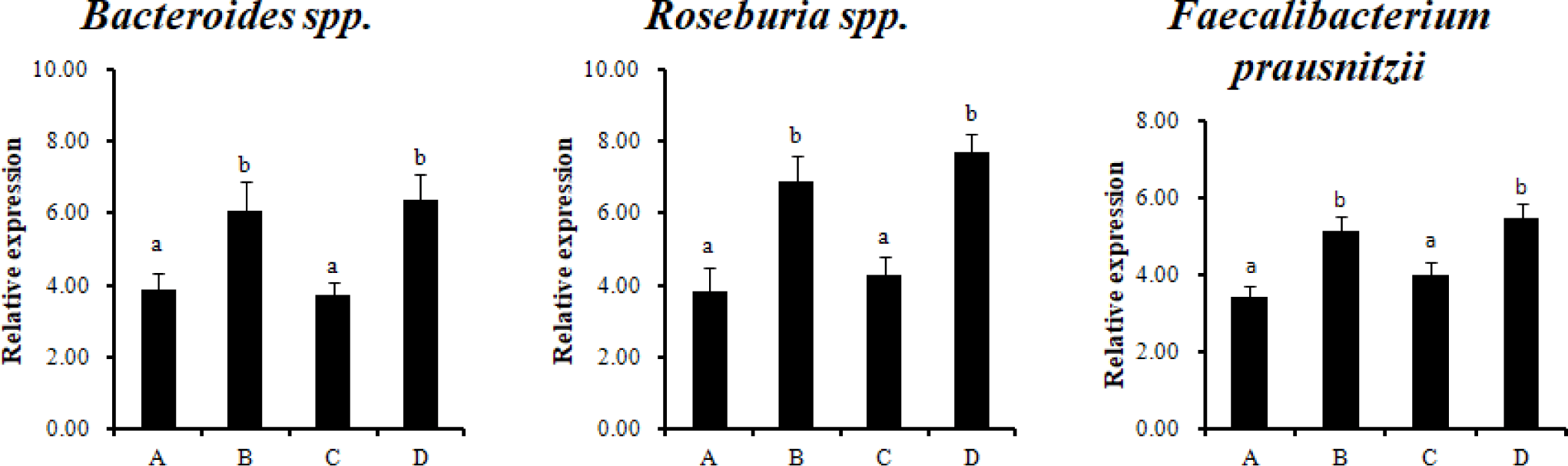
Analysis by qPCR using specific primers for twelve major types of intestinal microorganisms was performed to analyze the effect of RC-LAB fermented feed with probiotics on gut bacteria. Bacteroides spp., Roseburia spp., and Faecalibacterium prausnitzii (F. prausnitzii) were significantly higher in the treatment groups (B, D), which were fed RC-LAB fermented feed (p < 0.05) (Fig. 1). Firmicutes, Bacteroidetes, and Proteobacteria dominated the porcine intestinal microbiota at the phylum level.
The difference in the body weight of the pigs can be explained as a difference in the intestinal microbiota. For example, pigs with better feed efficiency tend to have a higher total volatile fatty acids concentration in the cecum and a higher butyric acid concentration in the colon, which can be explained as a difference in microbial composition and function.
Bacteroidetes spp., Roseburia species (belonging to the phylum Firmicutes, class Clostridia, order Clostridiales, and family Lachnospiraceae), and F. prausnitzii (phylum Firmicutes, Clostridium cluster IV) produce SCFAs (acetate, propionate, butyrate) using complex polysaccharides. In particular, the SCFA, butyrate, is used as an energy source for colonocytes. F. prausnitzii is the most abundant bacteria in healthy human and animal microbial communities [26]. It has an anti-inflammatory effect, so it is used as a potent biomarker for the diagnosis and prognosis of gut diseases [27]. Probiotics are mainly based on restoring the intestinal ecosystem balance naturally. Therefore, using the intestinal dominant commensal bacteria as next-generation probiotics (NGPs) is the most natural way to restore microbial imbalance in GIT. An alternative would be to develop fecal microbiota transplantation (FMT) strains and probiotics, parabiotics, and postbiotics for F. prausnitzii, one of the candidates for NGPs.
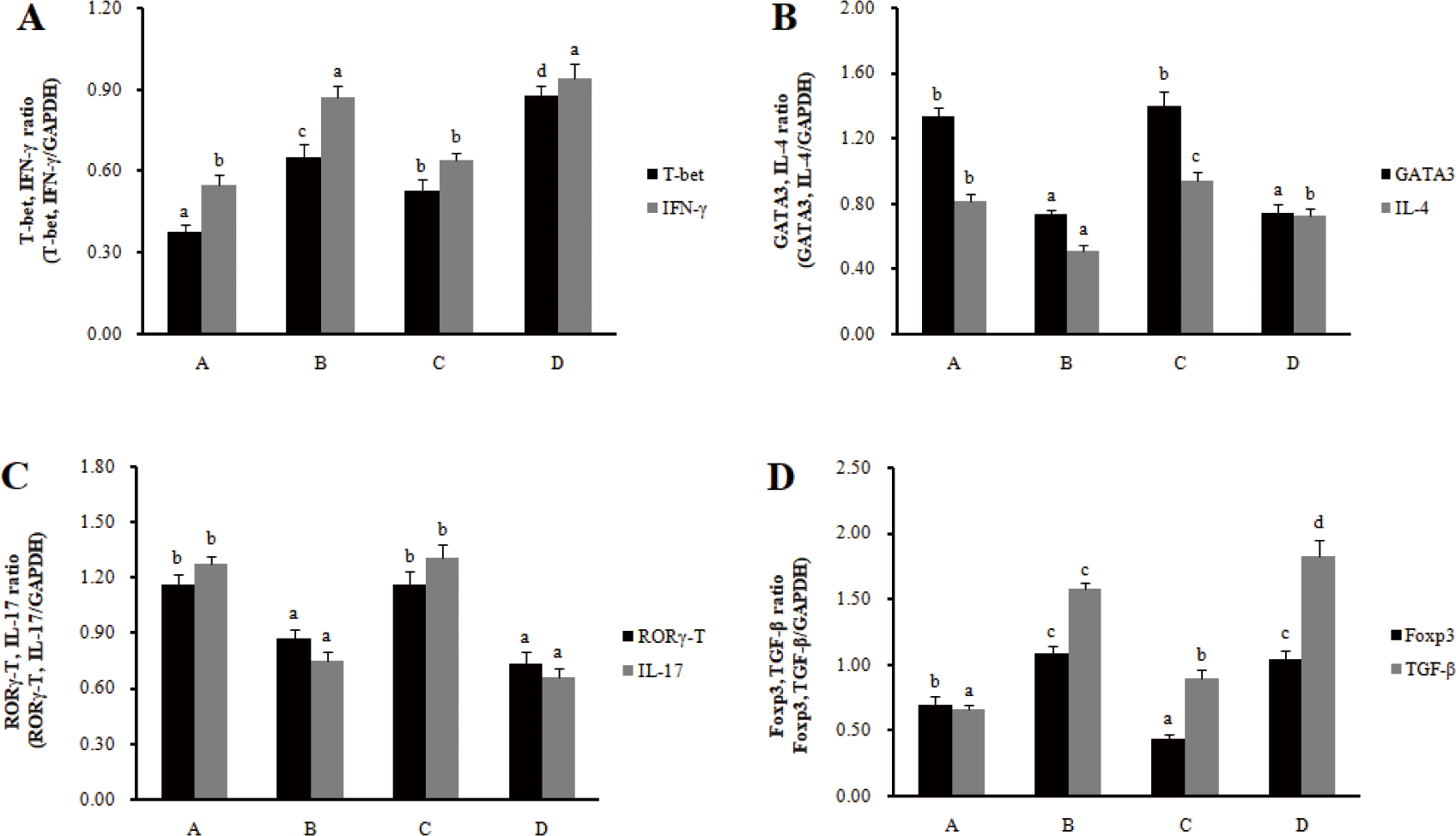
The mRNA expression of transcription factors (T-bet for Th1 cells, GATA 3 for Th2 cells, RORγT for Th17 cells, and Foxp3 for Treg cells) and cytokines (IFN-γ for Th1 cells, IL-4 for Th2 cells, IL-17 for Th17 cells, and IL-10 for Treg cells) was investigated to determine the effect of RC-LAB fermented feed on T cell polarization in MLN and spleens. In MLN, the mRNA expression of the transcription factors T-bet and cytokine IFN-γ in Th1 cells significantly increased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05), and the mRNA expression of the transcription factors GATA-3 and cytokine IL-4 in Th2 cells significantly decreased in treatment groups (B, D) compared to control groups (A, C) (p < 0.05) (Fig. 2). The mRNA expression of the transcription factors RORγ-T and cytokine IL-17 in Th17 cells significantly decreased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05), and the mRNA expression of the transcription factors Foxp3 and cytokine IL-10 in Treg cells significantly increased in the treatment groups (B, D) compared to the control groups (A, C) (p < 0.05).
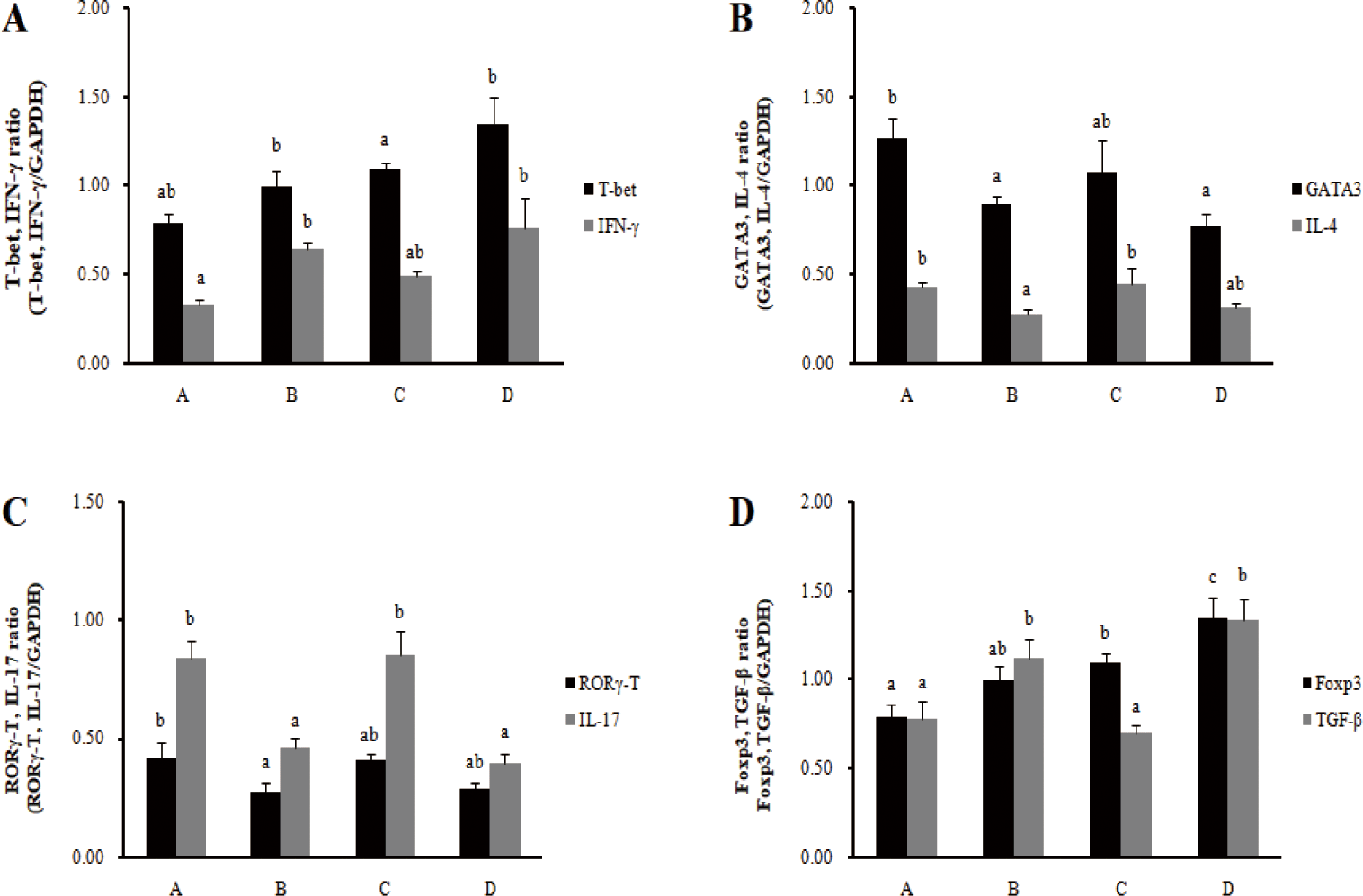
In the spleens, the mRNA expression of cytokine IFN-γ in Th1 cells increased significantly in the barrows treatment group (B), and the mRNA expression of transcription factor T-bet increased significantly in the gilts treatment group (D) (p < 0.05) (Fig. 3). The mRNA expression of transcription factors GATA-3 and cytokine IL-4 in Th2 cells significantly decreased in the barrows treatment group (B) (p < 0.05), and there was no significant difference in the gilts. In addition, the mRNA expression of the transcription factors RORγ-T and cytokine IL-17 in Th17 cells significantly decreased in the barrows treatment group (B) (p < 0.05), and cytokine IL-17 mRNA expression significantly decreased in the gilts (p < 0.05). For the mRNA expression of transcription factors Foxp3 and cytokine IL-10 in Treg cells, cytokine IL-10 significantly increased in the barrows treatment group (B) (p < 0.05). In the gilts treatment group (D), the mRNA expression of transcription factors Foxp3 and cytokine IL-10 significantly increased (p < 0.05). Feeding the RC-LAB fermented diet increased the transcription factors and cytokine transcription levels of Th1 cells and Treg cells in MLN, while decreasing the transcription levels of cytokines and the transcription factors of Th2 and Th17 cells. However, the immune modulatory function of Th1/Th2 and Th17/Treg in the spleens was less pronounced than in MLN.
In atopic dermatitis (AD)-induced mice treated with Duolac adenosine triphospha (ATP) (L. casei, L. plantarum, L. rhamnosus, and B. lactis), the Th1-mediated response upregulated downstream signaling molecules T-bet, STAT-1, and STAT-4 as well as IL-2 and IFN-gamma. Conversely, Duolac ATP inhibited Th2 and Th17 responses by downregulating GATA-3, C-maf, IL-4, IL-5, and IL-17. These results suggest that Duolac ATP regulates dendritic cells to initiate Th1 and Treg reactions [28]. In AD-induced mice, YK4, a probiotic mixture (Lactobacillus acidophilus CBT LA1, L. plantarum CBT LP3, Bifidobacterium breve CBT BR3, and B. lactis CBT BL3), induced the ratio of Th1 cells in spleens and the ratio of Treg cells in Peyer’s patches and MLN to inhibit the Th2 cell population, and Galectin-9 partially contributed to Tregs’ proliferation [29]. As a result of feeding sodium butyrate and a probiotic mixture (B. lactis, L. casei, L. rhamnosus, and L. plantarum) to AD-induced mice, allergic reactions were reduced by increasing Th1 and Treg cell differentiation in MLN and spleens [30]. In addition, in 16 month old mice, L. rhamnosus supplementation demonstrated the potential to increase healthy aging by mitigating the immunosenescence-associated Th1/Th2 imbalance and enhancing resistance to E. coli infection [31].
CONCLUSION
RC-LAB fermented feed with probiotics affected the intestinal microbiota composition of pigs, increasing beneficial and SCFAs-producing bacteria and decreasing harmful bacteria. It also modulated the balance of Th1/Th2 and Th17/Treg cells in intestinal lymph nodes and the spleen. LAB fermented feed with probiotics affected the intestinal microbiota composition of pigs and showed an effect of modulating intestinal immune homeostasis.
