RESEARCH ARTICLE
Genome-wide association studies on collagen contents trait for meat quality in Hanwoo
KyeongHye Won
1,#
, Dohyun Kim
1,#
, Inho Hwang
3
, Hak-Kyo Lee
1,2
, Jae-Don Oh
1,*
1Department of Animal Biotechnology, College of Agricultural and Life Sciences, Jeonbuk National University, Jeonju 54896, Korea
2Department of Agricultural Convergence Technology, Jeonbuk National University, Jeonju 54896, Korea
3Department of Animal Science, Jeonbuk National University, Jeonju 54896, Korea
# These authors contributed equally to this work.
*Corresponding author: Jae-Don Oh, Department of Animal Biotechnology, College of Agricultural and Life Sciences, Jeonbuk National University, Jeonju 54896, Korea. Tel: +82-63-270-5931, E-mail:
oh5ow@naver.com
© Copyright 2023 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: Oct 26, 2022; Revised: Nov 17, 2022; Accepted: Nov 19, 2022
Published Online: Mar 31, 2023
Abstract
Beef consumers valued meat quality traits such as texture, tenderness, juiciness, flavor, and meat color that determining consumers’ purchasing decision. Most research on meat quality has focused on marbling, a key characteristic related to meat eating quality. However, other important traits such as meat texture, tenderness, and color have not much studied in cattle. Among these traits, meat tenderness and texture of cattle are among the most important factors affecting quality evaluation of consumers. Collagen is the main component of connective tissues.It greatly affects meat tenderness. The objective of this study was to determine significant variants and candidate genes associated with collagen contents trait (total collagen) through genome-wide association studies (GWAS). Phenotypic and genomic data from 135 Hanwoo were used. The BLUPF90 family program and GRAMMAR method for GWAS were applied in this study. A total of 73 potential single nucleotide polymorphisms (SNPs) showed significant associations with collagen content. They were located in or near 108 candidate genes. TMEM135 and ME3 genes were identified to have the most significant SNPs associated with collagen contents trait. Data indicated that these genes were related to collagen. Biological processes and pathways for the prediction of biological functions of candidate genes were confirmed. We found that candidate genes were involved in positive regulation of CREB transcription factor activity and actin cytoskeleton related to tenderness and texture of beef. Three genes (CRTC3, MYO1C and MYLK4) belonging to these biological functions were related to tenderness. These results provide a basis for improving genomic characteristics of Hanwoo for the production of tender beef. Furthermore, they could be used they could be used as an index to select desired traits for consumers.
Keywords: Genome-wide association studies (GWAS); Single nucleotide polymorphisms (SNPs); Collagen; Meat quality; Tenderness; Hanwoo
INTRODUCTION
Korean beef consumers prefer Hanwoo cattle meat because of its tenderness and excellent flavor [1,2]. In addition, consumers have considered meat quality such as tenderness, texture, juiciness, flavor and meat color as important factors affecting their purchasing decisions. Recently, consumers prefer tender meat to fatty meat. Among various meat traits, tenderness and texture of cattle are among the most important factors affecting the quality evaluation of consumers [3–5].
Most beef researches have focused on increasing marbling related to fat, a key characteristic associated with meat eating quality. On the other hand, tenderness in cattle has not been much studied. Increasing meat quality by increasing fat consumes a lot of production costs due to long feeding periods and expensive feed costs. This causes production of a large amount of methane gas that contributes to global warming, which is also related to the environment. Therefore, it is necessary to produce tender beef with a short-term of breeding.
Collagen is an abundant connective tissue protein. It is a contributing factor to meat tenderness and texture. Collagen also plays important roles in quality of cooked meat. Collagen fibers shrink when heated, resulting in loss of fluid and less tender meat. They also serve to hold muscle fibers together after contraction. Post-mortem degradation of collagen and the use of collagenases appear to play a role in providing desired tenderness and texture by altering connective tissue structure. Collagen is very important for maintaining an acceptable texture [6–8].
Meat sensory characteristics such as tenderness, flavor, juiciness, and color are important meat quality parameters affected by biological characteristics and proteolytic activities of muscles. Biological characteristics of muscles such as collagen, fiber type, and intramuscular adipose tissue can regulate meat tenderness and flavor. They are known to be influenced by genetic and nurturing factors [9–12].
Advances in genotyping technologies have made it possible to identify many single nucleotide polymorphisms (SNPs) distributed throughout the whole genome. This further deepens the search for genomic insights into complex traits [13]. Genome-wide association studies (GWAS) enables the detection of specific markers, genomic regions, and candidate genes associated with economically important traits. They have been conducted in livestock using high-density panels to enable large-scale genotyping [14].
Thus, the objective of this study was to detect significant variants and candidate genes associated with collagen contents trait (total collagen) using GWAS. Furthermore, this study will ultimately contribute to the the production of tender beef with short feeding periods.
MATERIALS AND METHODS
Animals and phenotypic data
A total of 135 cattle of the Hanwoo (steers, n = 103; bull, n = 5; and cow, n = 27) were used in this study. Hanwoo were raised in the same feeding condition and slaughtered in Jeollabuk-do Province. After slaughter, their longissimus dorsi (LD) muscles were sampled and cut into 2.5 cm thick steaks. These muscle samples were vacuum-packed and stored at 4°C until 41 days postmortem [15].
Total collagen contents in each the sample was determined using the colorimetric method of Kolar [16] with suitable modifications. Briefly, 2 g of each sample was hydrolyzed with 7N H2SO4 at 105°C for 16 h. The hydrolysate was diluted with distilled water to 500 mL and filtered. Filter a part of the mixture into 100 mL Erlenmeyer flask, the filtrate is stable at least 2 weeks at 4°C. About 2 mL of diluted filtrate was taken and added with chloramine T solution into a tube and left at room temperature for 20 min. Thereafter, a 4-dimethylamino benzaldehyde solution was added and the mixture was heated at 60°C for 15 min. Absorbance values of samples and hydroxyproline standard were measured at 558 nm using a spectrophotometer. A standard calibration curve was plotted for 4-hydroxyproline and a regression line was drawn. Collagen content was expressed in mg/100 g sample after converting hydroxyproline into collagen with a conversion factor of 7.14. For insoluble (or heat stable) collagen contents, homogenized samples in Ringer’s solution [17] were heated at 77°C for 70 min, followed by centrifugation. Residual fractions were hydrolyzed in 7N H2SO4 for 16 h at 105°C. The hydroxyproline content of the hydrolysate was determined after neutralization according to the procedure of Kolar. The soluble collagen content was calculated based on the difference between total and insoluble collagen contents [18].
Genotypic data and quality control
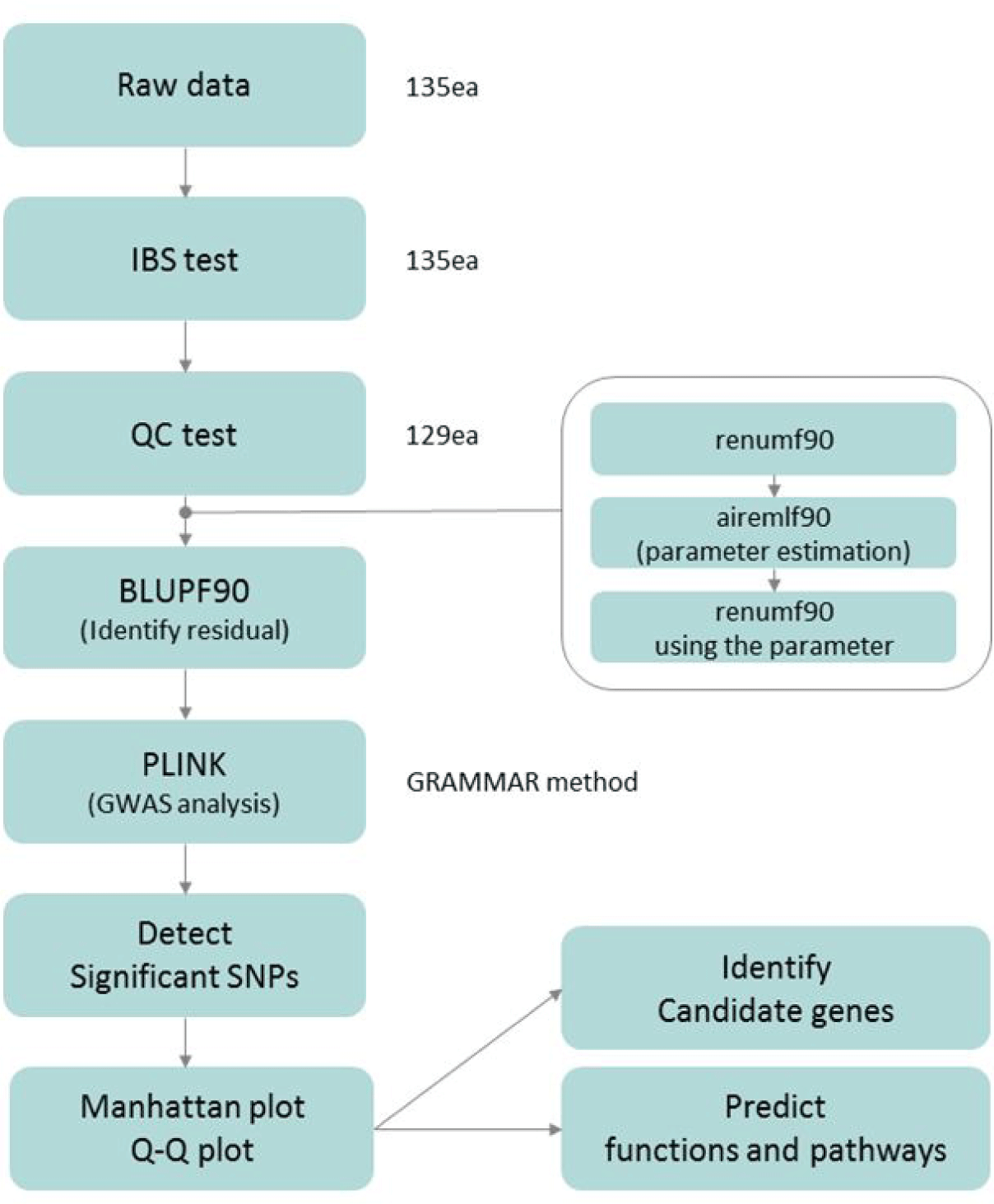
A total of 135 Hanwoo, consisting of steers (n = 103), bulls (n = 5), and cows (n = 27) were genotyped using a Bovine SNP50k chip (Illumina, San Diego, CA, USA). A total of 52,195 SNPs were collected. We performed the process of quality control (QC) based on the following criteria to ensure the quality of genotypic data obtained: i) removing individuals with identical genotype using identity by state (IBS) distance test (> 0.99), ii) eliminating individuals with call rates less than 90%, iii) removing SNPs with minor allele frequency less than 1%, iv) filtering out SNPs with call rates less than 90%, and v) removing SNPs with significant departure from Hardy Weinberg equilibrium (p < 10−7). These procedures were implemented with PLINK v1.07 [19] (Fig. 1).
Fig. 1.
Flow chart related to GWAS analysis using genomic and phenotypic data.
IBS, identity by state; QC, quality control; GWAS, genome-wide association studies; GWAS, genome-wide association studies; SNPs, single nucleotide polymorphisms; QQ, quantile-quantile.
Download Original Figure
Statistical analysis
BLUPF90 is, a software that comprises a family of program in Fortran 90/95 for mixed model computations in animal breeding [20]. It was used to estimate variance components and genetic parameters of collagen contents trait and residuals that were difference between the actual phenotype value and the estimated value in order to identify only genetic effects. First, QC data were renumbered and variance was estimated using RENUMF90. Second, we estimated variance components and genetic parameter using AIREMLF90 [21]. Third, we performed RENUMF90 analysis one more time to improve the accuracy of analysis using the estimated variance components and genetic parameters. Finally, these various components were then used to identify residuals. These procedures were performed using a multiple-trait model. Using the multiple-trait model, we estimated the variance component and genetic parameters of the collagen contents traits. The equation was as follows:
Where Y was the vector of phenotypic observations for total collagen, heat insoluble collagen, or soluble collagen contents; β was the vector of fixed effects including contemporary group effects, time of slaughtering (year, month), time of age at slaughter (month, days, age) and sex (cow, bull, steer) as a linear covariate; a was the vector of direct additive effects; e was the vector of residual random effects; X was the incidence matrix relating the phenotypes to the fixed effects; and Z was the incidence matrix relating the animal to the phenotype [22].
Using genome-wide rapid association using mixed model and regression (GRAMMAR) [23] in PLINK and the residuals obtained, we estimated SNP effect that affected collagen contents trait in the meat of Hanwoo (Fig. 1).
Identification of significant SNPs and annotation of candidate genes
We obtained significant SNPs associated with the phenotype based on p- < 0.001. Candidate genes associated with significant SNPs were annotated within 500 kb downstream and upstream of detected SNPs based on the Bos taurus transfer format (GTF) (version ARS-UCD1.2.104) in Ensembl database.
Functional analysis
We performed Gene Ontology (GO) [24] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [25] pathway analyses to investigate functions of candidate genes using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [26].
RESULTS
Phenotypes and genotypes
We identified 129 individuals after removing six (ID: 002311515862, 002311652670, 002300057089, 002085007947, 002114973907 and 002112336318) out of 135 individuals by IBS distance test and outlier. We identified basic statistics for phenotypic data of 129 Hanwoo after QC. The estimates for collagen contents traits were on average 0.362, 0.250 and 0.114 respectively (Table 1).
Table 1.
The basic statistics for phenotypic data
|
Traits |
N |
Min |
Max |
Mean |
SD |
|
Total collagen (TC) |
129 |
0.140 |
1.500 |
0.362 |
0.216 |
|
Heat insoluble collagen (HC) |
129 |
0.030 |
1.310 |
0.250 |
0.194 |
|
Soluble collagen (SC) |
129 |
−0.020 |
0.310 |
0.114 |
0.069 |
Download Excel Table
Through the QC test, 5,715 SNPs out of 52,195 SNPs were removed and 46,480 SNPs associated with collagen contents trait were finally used for GWAS analysis. We confirmed the difference in the number of available SNPs per chromosome and the interval size (kb) of each autosome of autosuomes 1 to 29 before and after QC. We identified the number of SNPs considered useful for Hanwoo per chromosome, ranging from a minimum of 799 to a maximum of 2,909. About 89.1% of total SNPs were selected as available SNPs. The average distance of adjacent SNPs for each chromosome was 54.343 kb (Table 2).
Table 2.
The number of available SNPs and average interval distance between adjacent SNPs in Bovine SNP50k chip
|
BTA |
Number of SNPs |
Remove frequency (%) |
Mean of Interval SNP |
|
Before QC |
After QC |
Before QC |
After QC |
|
1 |
3,225 |
2,909 |
0.902 |
49.037 |
54.365 |
|
2 |
2,756 |
2,444 |
0.887 |
49.614 |
55.94 |
|
3 |
2,582 |
2,280 |
0.883 |
46.937 |
53.157 |
|
4 |
2,479 |
2,214 |
0.893 |
48.678 |
54.507 |
|
5 |
2,156 |
1,912 |
0.887 |
56.185 |
63.359 |
|
6 |
3,158 |
2,820 |
0.893 |
37.698 |
42.218 |
|
7 |
2,481 |
2,250 |
0.907 |
45.316 |
49.971 |
|
8 |
2,246 |
2,011 |
0.895 |
50.339 |
56.224 |
|
9 |
2,077 |
1,858 |
0.895 |
50.802 |
56.793 |
|
10 |
2,357 |
2,085 |
0.885 |
44.216 |
49.987 |
|
11 |
2,181 |
1,915 |
0.878 |
49.164 |
55.997 |
|
12 |
1,651 |
1,427 |
0.864 |
55.118 |
63.776 |
|
13 |
1,684 |
1,521 |
0.903 |
49.829 |
55.173 |
|
14 |
2,274 |
1,965 |
0.864 |
36.583 |
42.338 |
|
15 |
1,681 |
1,486 |
0.884 |
50.437 |
57.06 |
|
16 |
1,599 |
1,420 |
0.888 |
50.947 |
57.374 |
|
17 |
1,567 |
1,386 |
0.884 |
47.84 |
54.07 |
|
18 |
1,303 |
1,167 |
0.896 |
50.231 |
55.883 |
|
19 |
1,380 |
1,243 |
0.901 |
46.078 |
51.16 |
|
20 |
1,571 |
1,388 |
0.884 |
45.602 |
51.618 |
|
21 |
1,398 |
1,255 |
0.898 |
50.893 |
56.697 |
|
22 |
1,212 |
1,085 |
0.895 |
50.55 |
56.472 |
|
23 |
1,124 |
1,019 |
0.907 |
46.505 |
51.175 |
|
24 |
1,230 |
1,108 |
0.901 |
50.745 |
56.137 |
|
25 |
939 |
847 |
0.902 |
45.633 |
50.596 |
|
26 |
1,032 |
922 |
0.893 |
49.511 |
55.324 |
|
27 |
918 |
823 |
0.897 |
49.435 |
55.149 |
|
28 |
905 |
799 |
0.883 |
51.126 |
57.873 |
|
29 |
1,029 |
921 |
0.895 |
50.084 |
55.546 |
|
Total |
52,195 |
46,480 |
0.891 |
48.453 |
54.343 |
Download Excel Table
GWAS analysis
We identified residuals using BLUPF90 for GWAS analysis. These residuals were differences between observed and estimated phenotypic values. In the total collagen contents trait, the observed value was 0.180 and the estimated value was 0.594. Thus, the residual value was the lowest at −0.414 (ID: 002083966119). Observed and estimated values were 1.500 and 0.625, respectively. The residual value was the highest at 0.875 (ID: 002083963503) (Table 3).
Table 3.
The basic statistics for results of BLUPF90 analysis
|
Statistics |
N |
Min |
Max |
Range |
Mean |
Median |
SE |
VAR |
SD |
Coef.var |
|
TC_observation |
129 |
−0.209 |
1.5 |
1.709 |
0.359 |
0.31 |
0.019 |
0.049 |
0.221 |
0.615 |
|
TC_estimation |
129 |
0.059 |
0.696 |
0.637 |
0.361 |
0.344 |
0.012 |
0.019 |
0.139 |
0.385 |
|
TC_residual |
129 |
−0.414 |
0.875 |
1.289 |
0 |
−0.015 |
0.015 |
0.027 |
0.165 |
2.05E+17 |
Download Excel Table
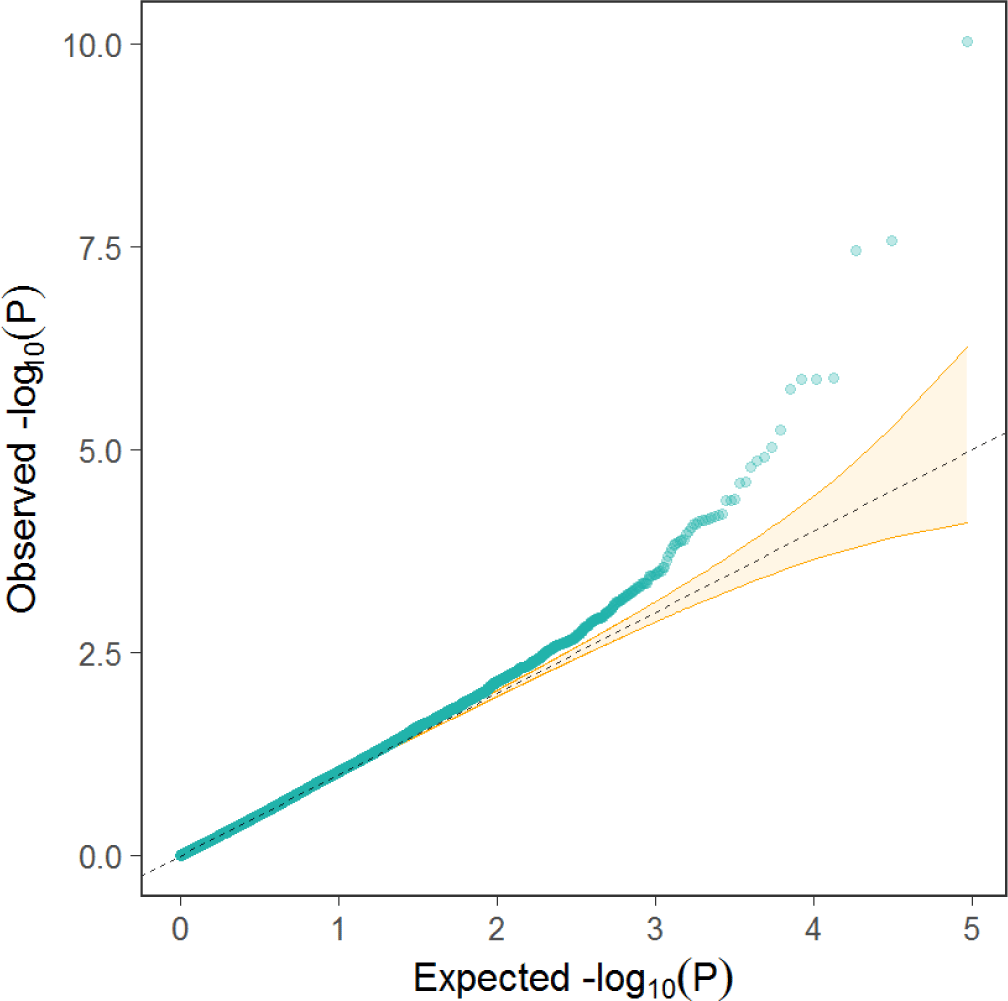
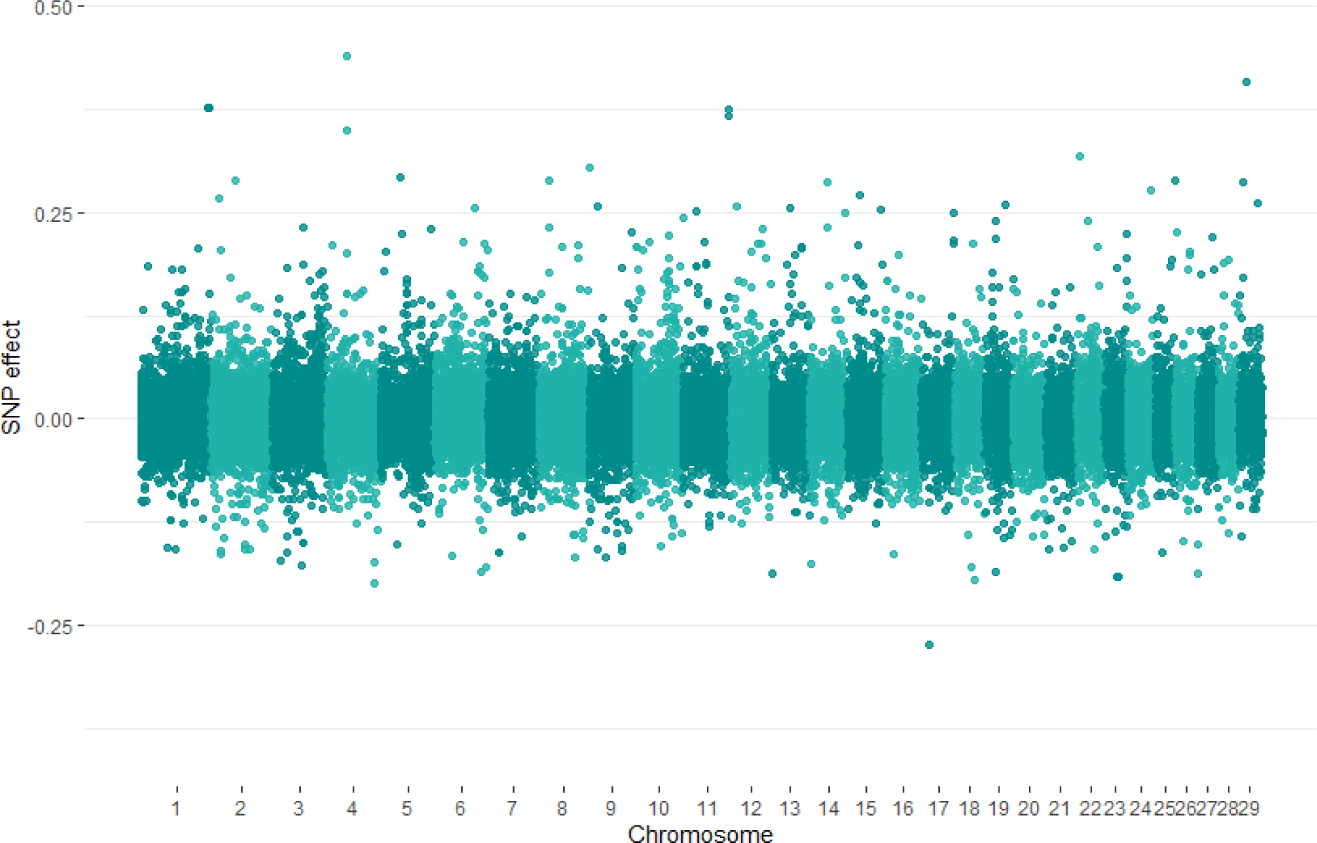
The quantile-quantile (QQ) plot and inflation control (lambda) value were used to compare observed distributions of −log10(p) to the expected distribution under the no association model for total collagen contents trait (Fig. 2). The QQ plot for trait showed that the mixed linear model fitted the data well. Estimates of SNP effects associated with total collagen contents trait were within a range of −0.273 to 0.438 (Fig. 3).
Fig. 2.
The QQ-plot for the studied total collagen contents trait.
The dotted line represents the 95% concentration band under the null hypothesis no association between trait and SNPs. The green dots represent the p-values. QQ, quantile-quantile; SNPs, single nucleotide polymorphisms.
Download Original Figure
Fig. 3.
The Manhattan plot of SNP effects for GWAS analysis using GRAMMAR methods in PLINK.
SNP, single nucleotide polymorphism; GWAS, genome-wide association studies.
Download Original Figure
Identification of significant SNPs and annotation of candidate genes
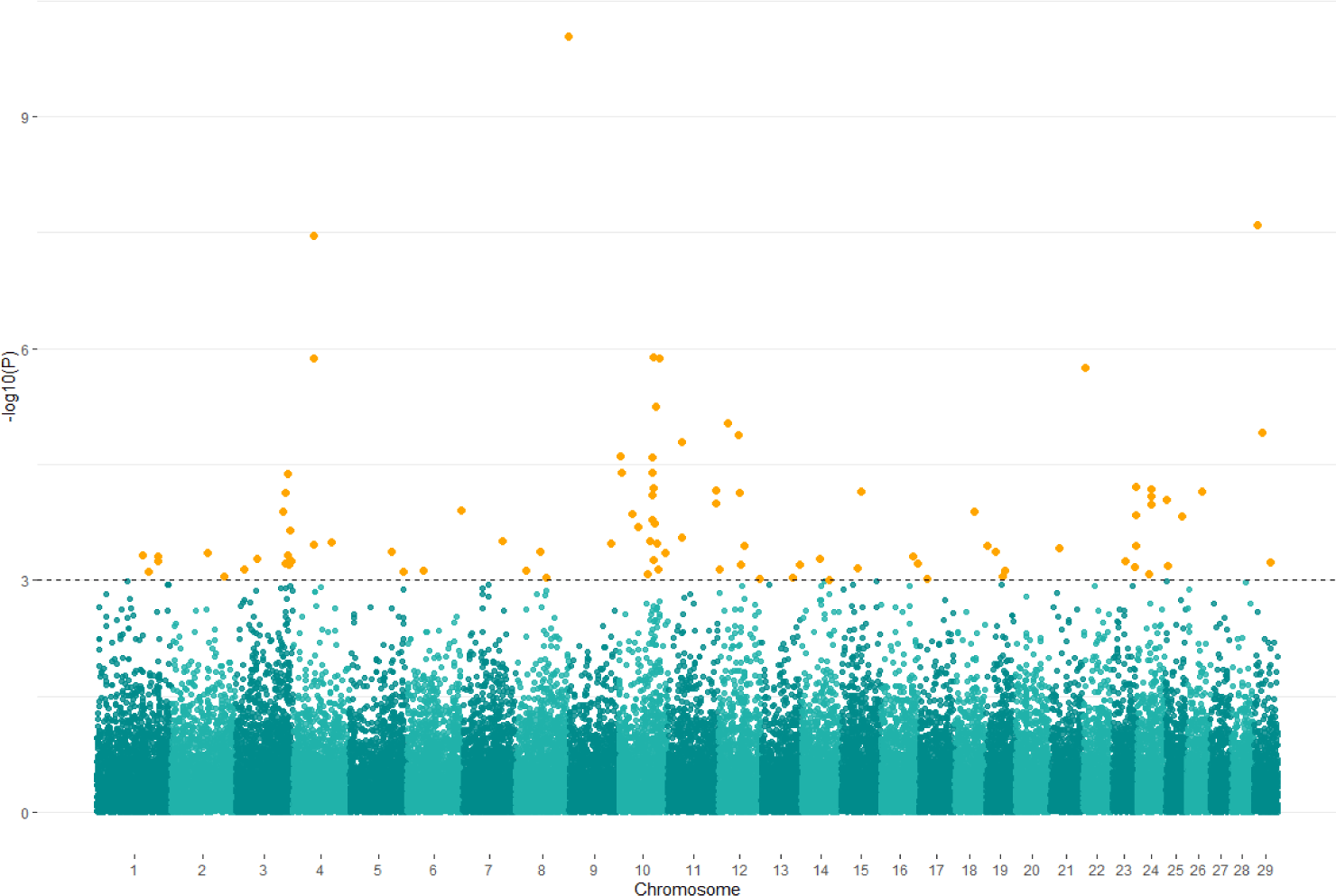
We prepared a Manhattan plot of −log10(p) for genomic positions of SNP markers using p-value from the SNP effect result to confirm the significant SNP associated with total collagen contents trait. Based on p < 0.001, a total of 73 SNPs were significantly associated with the trait (Fig. 4 and Table 4). A total of 108 candidate genes associated with significant SNPs were annotated based on Bos taurus genome. The most significant SNP was confirmed to be UA-IFASA-8514 (chr29:8936383) in Transmembrane protein 135 (TMEM135) and Malic Enzyme 3 (ME3) genes.
Table 4.
Significant SNPs (total collagen)
|
SNP_id |
SNP (MinorA/ MajorB) |
Chr |
Genomic position |
Estimate |
SE |
p-value |
−LogP |
nAA |
nAB |
nBB |
σ2 (SNP)/ σ2 (trait) |
Gene_name |
|
Hapmap38956-BTA-43309 |
[C/A] |
1 |
97,366,225 |
0.1576 |
0.0440 |
0.0005 |
3.3186 |
1 |
10 |
119 |
0.0804 |
SLC7A14 |
|
ARS-BFGL-NGS-535 |
[G/A] |
1 |
109,221,369 |
−0.0636 |
0.0185 |
0.0008 |
3.1119 |
27 |
52 |
51 |
0.0719 |
MFSD1, RSRC1 |
|
rs383007308 |
[A/G] |
1 |
131,149,414 |
0.1204 |
0.0340 |
0.0006 |
3.2490 |
1 |
22 |
107 |
0.0893 |
ESYT3 |
|
ARS-BFGL-NGS-109828 |
[G/A] |
2 |
77,033,601 |
0.1496 |
0.0415 |
0.0004 |
3.3557 |
1 |
13 |
116 |
0.0894 |
CNTNAP5 |
|
rs110414144 |
[A/C] |
2 |
113,584,292 |
0.0834 |
0.0245 |
0.0009 |
3.0467 |
5 |
48 |
77 |
0.0886 |
DOCK10, NYAP2 |
|
3:101365765 |
[C/A] |
3 |
101,365,765 |
0.1330 |
0.0337 |
0.0001 |
3.8871 |
1 |
22 |
107 |
0.1090 |
ZSWIM5, TMEM53 |
|
rs456169856 |
[A/G] |
3 |
106,866,376 |
0.1053 |
0.0299 |
0.0006 |
3.2230 |
3 |
22 |
105 |
0.0783 |
MACF1 |
|
BTB-01834875 |
[G/A] |
3 |
46,721,234 |
−0.0763 |
0.0215 |
0.0005 |
3.2758 |
11 |
47 |
72 |
0.0834 |
DPYD, PTBP2 |
|
ARS-BFGL-NGS-13158 |
[C/G] |
3 |
19,337,220 |
0.0790 |
0.0228 |
0.0007 |
3.1439 |
8 |
48 |
74 |
0.0851 |
POGZ |
|
BTB-00157833 |
[G/A] |
3 |
111,842,741 |
0.0711 |
0.0198 |
0.0005 |
3.3249 |
19 |
57 |
54 |
0.0861 |
CSMD2 |
|
rs381136948 |
[A/C] |
3 |
112,012,156 |
0.1779 |
0.0419 |
4.E-05 |
4.3832 |
1 |
11 |
118 |
0.1105 |
CSMD2, GIGYF2 |
|
Hapmap57254-rs29022776 |
[A/G] |
3 |
113,433,557 |
0.0763 |
0.0218 |
0.0006 |
3.1947 |
12 |
62 |
56 |
0.0947 |
MROH2A |
|
rs385230778 |
[A/G] |
3 |
115,009,419 |
0.1587 |
0.0419 |
0.0002 |
3.6379 |
2 |
7 |
121 |
0.0750 |
AGAP1 |
|
ARS-BFGL-NGS-118221 |
[A/C] |
5 |
90,684,739 |
0.0998 |
0.0276 |
0.0004 |
3.3663 |
2 |
40 |
88 |
0.1028 |
PLEKHA5 |
|
Hapmap41631-BTA-75177 |
[A/G] |
5 |
114,405,063 |
0.0685 |
0.0199 |
0.0008 |
3.1054 |
16 |
45 |
69 |
0.0720 |
EFCAB6 |
|
ARS-BFGL-NGS-11339 |
[A/G] |
4 |
45,128,142 |
0.3496 |
0.0689 |
1.E-06 |
5.8765 |
0 |
5 |
125 |
0.1695 |
RELN, LHFPL3 |
|
BTB-00182993 |
[C/A] |
4 |
45,513,003 |
0.4383 |
0.0746 |
3.E-08 |
7.4631 |
0 |
4 |
126 |
0.2139 |
RELN, LHFPL3 |
|
BTA-70441-no-rs |
[G/A] |
4 |
45,602,047 |
0.2001 |
0.0544 |
0.0003 |
3.4599 |
0 |
9 |
121 |
0.0984 |
RELN, LHFPL3, ENSBTAG00000048818 |
|
ARS-BFGL-NGS-110196 |
[G/A] |
4 |
81,109,721 |
−0.0970 |
0.0262 |
0.0003 |
3.4935 |
4 |
33 |
93 |
0.0919 |
SUGCT, POU6F2 |
|
Hapmap23877-BTA-143906 |
[A/C] |
6 |
36,829,725 |
0.1503 |
0.0435 |
0.0007 |
3.1284 |
0 |
16 |
114 |
0.0959 |
HERC6, NCAPG |
|
BTB-01265106 |
[A/C] |
6 |
116,779,750 |
0.2034 |
0.0514 |
0.0001 |
3.8972 |
0 |
11 |
119 |
0.1232 |
ZFYVE28, FGFR3 |
|
BTB-00348139 |
[A/G] |
8 |
52,645,428 |
0.1334 |
0.0369 |
0.0004 |
3.3626 |
0 |
22 |
108 |
0.1013 |
PCSK5 |
|
ARS-BFGL-NGS-66538 |
[A/G] |
8 |
64,011,227 |
−0.1006 |
0.0297 |
0.0009 |
3.0301 |
2 |
29 |
99 |
0.0825 |
GABBR2, COL15A1 |
|
ARS-BFGL-NGS-34771 |
[A/G] |
8 |
21,943,761 |
0.1775 |
0.0513 |
0.0007 |
3.1288 |
1 |
6 |
123 |
0.0691 |
ENSBTAG00000053368 |
|
BTB-00960162 |
[A/G] |
7 |
83,915,853 |
0.1482 |
0.0400 |
0.0003 |
3.5092 |
0 |
18 |
112 |
0.1040 |
VCAN, EDIL3 |
|
rs443739156 |
[C/A] |
11 |
30,840,950 |
0.1851 |
0.0495 |
0.0003 |
3.5511 |
0 |
11 |
119 |
0.1021 |
STON1, FSHR |
|
ARS-BFGL-NGS-83866 |
[A/G] |
11 |
30,851,124 |
0.2519 |
0.0562 |
2.E-05 |
4.7918 |
0 |
8 |
122 |
0.1391 |
STON1, FSHR |
|
BTB-01944534 |
[A/G] |
11 |
30,826,527 |
0.1851 |
0.0495 |
0.0003 |
3.5511 |
0 |
11 |
119 |
0.1021 |
FSHR |
|
ARS-BFGL-NGS-2573 |
[A/G] |
11 |
101,602,103 |
0.3742 |
0.0908 |
0.0001 |
4.1669 |
0 |
3 |
127 |
0.1174 |
ABL1, PRRC2B, MED27 |
|
ARS-BFGL-NGS-94862 |
[G/A] |
11 |
103,534,103 |
0.3660 |
0.0911 |
0.0001 |
3.9978 |
0 |
3 |
127 |
0.1123 |
NACC2 |
|
BTA-62308-no-rs |
[G/A] |
10 |
29,176,267 |
0.2134 |
0.0542 |
0.0001 |
3.8626 |
0 |
9 |
120 |
0.1127 |
AVEN, FMN1 |
|
ARS-BFGL-NGS-3005 |
[C/A] |
10 |
81,981,544 |
0.1212 |
0.0329 |
0.0003 |
3.4729 |
2 |
19 |
109 |
0.0871 |
SYNJ2BP |
|
BTA-60292-no-rs |
[A/G] |
10 |
7,308,238 |
0.1575 |
0.0370 |
4.E-05 |
4.3893 |
1 |
16 |
113 |
0.1175 |
CERT1, IQGAP2 |
|
ARS-BFGL-NGS-118392 |
[G/A] |
10 |
84,632,536 |
0.1263 |
0.0364 |
0.0007 |
3.1479 |
1 |
18 |
111 |
0.0833 |
DPF3, PSEN1, MIDEAS |
|
Hapmap41972-BTA-79298 |
[A/G] |
10 |
85,547,284 |
0.1577 |
0.0311 |
1.E-06 |
5.8690 |
3 |
15 |
112 |
0.1357 |
LIN52, YLPM1 |
|
Hapmap39952-BTA-86345 |
[C/A] |
10 |
99,626,471 |
0.0730 |
0.0202 |
0.0004 |
3.3565 |
28 |
69 |
33 |
0.0978 |
SPATA7 |
|
Hapmap44561-BTA-72345 |
[G/A] |
10 |
60,168,183 |
0.1684 |
0.0492 |
0.0008 |
3.0779 |
1 |
7 |
122 |
0.0697 |
TRPM7, ATP8B4 |
|
ARS-BFGL-NGS-110504 |
[A/C] |
10 |
70,556,929 |
0.1485 |
0.0383 |
0.0002 |
3.7812 |
1 |
15 |
114 |
0.0991 |
PSMA3 |
|
ARS-BFGL-NGS-116012 |
[G/A] |
10 |
71,266,946 |
0.1678 |
0.0411 |
0.0001 |
4.0971 |
1 |
12 |
115 |
0.1070 |
KIAA0586, JKAMP |
|
BTB-00434096 |
[G/A] |
10 |
71,082,204 |
0.1779 |
0.0419 |
4.E-05 |
4.3862 |
1 |
11 |
118 |
0.1105 |
DAAM1 |
|
ARS-BFGL-NGS-23163 |
[A/G] |
10 |
71,835,260 |
0.1944 |
0.0445 |
3.E-05 |
4.5979 |
1 |
9 |
120 |
0.1126 |
RTN1, PCNX4 |
|
BTB-01203179 |
[A/G] |
10 |
72,694,329 |
0.0909 |
0.0220 |
0.0001 |
4.2010 |
9 |
52 |
69 |
0.1195 |
LRRC9, SLC38A6 |
|
ARS-BFGL-NGS-69839 |
[G/A] |
10 |
74,011,076 |
0.2210 |
0.0435 |
1.E-06 |
5.8897 |
1 |
9 |
120 |
0.1455 |
PRKCH, SYT16 |
|
rs378634523 |
[A/G] |
10 |
76,078,820 |
0.0913 |
0.0237 |
0.0002 |
3.7395 |
6 |
45 |
79 |
0.1048 |
GPHB5, MTHFD1 |
|
ARS-BFGL-NGS-116025 |
[T/A] |
10 |
79,496,312 |
0.1353 |
0.0286 |
6.E-06 |
5.2450 |
2 |
30 |
98 |
0.1530 |
GPHN, RAD51B |
|
ARS-BFGL-NGS-101050 |
[G/A] |
12 |
19,868,800 |
0.0917 |
0.0199 |
9.E-06 |
5.0292 |
21 |
67 |
42 |
0.1504 |
RNASEH2B |
|
BTA-21437-no-rs |
[C/A] |
12 |
43,601,825 |
0.0932 |
0.0205 |
1.E-05 |
4.8732 |
14 |
51 |
60 |
0.1379 |
KLHL1 |
|
Hapmap45915-BTA-22734 |
[A/T] |
12 |
46,867,626 |
0.0742 |
0.0212 |
0.0006 |
3.2022 |
12 |
48 |
70 |
0.0811 |
DACH1 |
|
Hapmap51092-BTA-93283 |
[A/G] |
12 |
3,172,154 |
−0.0684 |
0.0197 |
0.0007 |
3.1424 |
19 |
54 |
57 |
0.0786 |
DIAPH3 |
|
ARS-BFGL-NGS-43617 |
[A/G] |
15 |
35,138,494 |
0.1610 |
0.0463 |
0.0007 |
3.1627 |
0 |
13 |
117 |
0.0905 |
SERGEF, PLEKHA7 |
|
ARS-BFGL-NGS-118358 |
[G/A] |
15 |
42,960,757 |
0.1453 |
0.0353 |
0.0001 |
4.1554 |
0 |
24 |
105 |
0.1310 |
SBF2, SWAP70, DENND5A |
|
Hapmap40712-BTA-33406 |
[A/G] |
13 |
67,101,174 |
0.1238 |
0.0365 |
0.0009 |
3.0383 |
1 |
18 |
111 |
0.0800 |
KIAA1755, PPP1R16B |
|
ARS-BFGL-NGS-115847 |
[A/G] |
14 |
40,458,050 |
0.2866 |
0.0805 |
0.0005 |
3.2804 |
0 |
4 |
125 |
0.0922 |
ZFHX4 |
|
ARS-BFGL-NGS-116702 |
[A/G] |
16 |
79,720,573 |
0.1447 |
0.0411 |
0.0006 |
3.2188 |
0 |
17 |
113 |
0.0941 |
IGFN1, PPP1R12B |
|
BTA-40408-no-rs |
[G/C] |
17 |
17,408,409 |
−0.2725 |
0.0805 |
0.0010 |
3.0203 |
0 |
4 |
126 |
0.0827 |
RNF150, MAML3 |
|
ARS-BFGL-NGS-98331 |
[A/G] |
21 |
22,057,989 |
0.1534 |
0.0421 |
0.0004 |
3.4121 |
0 |
16 |
114 |
0.0999 |
CRTC3, SLC28A1 |
|
Hapmap42198-BTA-39980 |
[G/A] |
18 |
41,617,350 |
−0.0790 |
0.0200 |
0.0001 |
3.8824 |
30 |
68 |
32 |
0.1145 |
ZNF536 |
|
ARS-BFGL-NGS-111273 |
[A/G] |
19 |
37,699,961 |
−0.0877 |
0.0258 |
0.0009 |
3.0501 |
4 |
40 |
86 |
0.0850 |
PHB, CALCOCO2, SKAP1 |
|
Hapmap60163-rs29015084 |
[G/A] |
19 |
22,120,443 |
0.0910 |
0.0252 |
0.0004 |
3.3654 |
4 |
45 |
81 |
0.0988 |
MYO1C |
|
ARS-BFGL-NGS-32460 |
[G/A] |
19 |
40,716,234 |
0.0721 |
0.0209 |
0.0007 |
3.1266 |
14 |
56 |
60 |
0.0835 |
TNS4 |
|
BTB-00885986 |
[A/C] |
24 |
31,423,445 |
−0.0754 |
0.0188 |
0.0001 |
3.9739 |
31 |
60 |
39 |
0.1040 |
ZNF521 |
|
BTB-00885964 |
[C/A] |
24 |
31,457,933 |
−0.0771 |
0.0189 |
0.0001 |
4.0891 |
31 |
61 |
38 |
0.1088 |
ZNF521 |
|
Hapmap49083-BTA-21452 |
[A/G] |
24 |
32,171,355 |
0.0820 |
0.0199 |
0.0001 |
4.1832 |
16 |
53 |
61 |
0.1088 |
ZNF521 |
|
BTB-00830153 |
[A/G] |
22 |
4,534,118 |
0.3169 |
0.0633 |
2.E-06 |
5.7484 |
0 |
6 |
124 |
0.1665 |
RBMS3 |
|
ARS-BFGL-NGS-103489 |
[A/G] |
23 |
49,135,538 |
0.1671 |
0.0479 |
0.0007 |
3.1708 |
0 |
13 |
117 |
0.0975 |
FARS2, CDYL |
|
ARS-BFGL-NGS-27800 |
[A/G] |
23 |
50,897,089 |
0.1354 |
0.0369 |
0.0004 |
3.4454 |
1 |
17 |
112 |
0.0913 |
SLC22A23, MYLK4, GMDS |
|
ARS-BFGL-NGS-108494 |
[A/G] |
23 |
51,241,585 |
0.1951 |
0.0471 |
0.0001 |
4.2057 |
0 |
12 |
118 |
0.1232 |
SERPINB6, GMDS |
|
ARS-BFGL-NGS-13891 |
[A/G] |
23 |
51,487,543 |
0.2238 |
0.0571 |
0.0001 |
3.8431 |
0 |
8 |
122 |
0.1098 |
GMDS |
|
ARS-BFGL-NGS-27911 |
[A/G] |
26 |
35,048,102 |
0.1982 |
0.0483 |
0.0001 |
4.1410 |
1 |
7 |
122 |
0.0965 |
ABLIM1, ATRNL1 |
|
UA-IFASA-8514 |
[C/A] |
29 |
8,936,383 |
0.2864 |
0.0482 |
3.E-08 |
7.5884 |
0 |
11 |
119 |
0.2443 |
TMEM135, ME3 |
|
Hapmap27205-BTA-156398 |
[A/G] |
29 |
18,203,037 |
0.4084 |
0.0897 |
1.E-05 |
4.9151 |
0 |
3 |
127 |
0.1399 |
GAB2, RSF1 |
|
ARS-BFGL-NGS-92926 |
[C/A] |
25 |
4,131,112 |
0.1035 |
0.0296 |
0.0007 |
3.1795 |
3 |
23 |
104 |
0.0780 |
HMOX2, ENSBTAG00000026383 |
|
ARS-BFGL-BAC-42642 |
[A/G] |
25 |
35,532,374 |
0.1930 |
0.0493 |
0.0001 |
3.8300 |
0 |
12 |
118 |
0.1206 |
CUX1 |
Download Excel Table
Fig. 4.
The Manhattan plot of GWAS for total collagen contents trait with significance thresholds indicated at −log10P > 1 × 10−3.
The orange dots represent significant SNPs associated with total collagen contents trait. GWAS, genome-wide association studies; SNPs, single nucleotide polymorphisms.
Download Original Figure
Functional analysis
We identified functions of candidate genes associated with total collagen contents trait from GO and KEGG analysis using DAVID (Tables 5 and 6). In biological process from GO, we confirmed eight gene ontologies: negative regulation of peptidyl-serine dephosphorylation (GO:1902309), negative regulation of ubiquitin-protein transferase activity (GO:0051444), receptor localization to synapse (GO:0097120), T cell receptor signaling pathway (GO:0050852), negative regulation of epidermal growth factor-activated receptor activity (GO:0007175), somite development (GO:0061053), positive regulation of CREB transcription factor activity (GO:0032793), and amino acid transmembrane transport (GO:0003333). In cellular component, we identified seven gene ontologies: cytosol (GO:0005829), nucleoplasm (GO:0005654), actin cytoskeleton (GO:0015629), perinuclear region of cytoplasm (GO:0048471), cytoplasm (GO:0005737), nuclear speck (GO:0016607), and cell projection (GO:0042995). Through KEGG analysis, we identified regulation of actin cytoskeleton (bta04810) pathway.
Table 5.
The results of gene ontology (GO) analysis of candidate genes associated with total collagen contents trait from GWAS analysis
|
GO ID |
Description |
#Genes |
Fold enrichment |
p-value |
Gene name |
|
Biological process |
|
GO:1902309 |
Negative regulation of peptidyl-serine dephosphorylation |
2 |
181.846 |
0.011 |
PPP1R16B, SWAP70 |
|
GO:0051444 |
Negative regulation of ubiquitin-protein transferase activity |
2 |
51.956 |
0.037 |
ABL1, PSEN1 |
|
GO:0097120 |
Receptor localization to synapse |
2 |
40.410 |
0.048 |
RELN, SYNJ2BP |
|
GO:0050852 |
T cell receptor signaling pathway |
3 |
7.577 |
0.059 |
ABL1, PSEN1, SKAP1 |
|
GO:0007175 |
Negative regulation of epidermal growth factor-activated receptor activity |
2 |
30.308 |
0.063 |
ZFYVE28, PSEN1 |
|
GO:0061053 |
Somite development |
2 |
25.978 |
0.074 |
RAD51B, MTHFD1 |
|
GO:0032793 |
Positive regulation of CREB transcription factor activity |
2 |
24.246 |
0.079 |
CRTC3, RELN |
|
GO:0003333 |
Amino acid transmembrane transport |
2 |
20.205 |
0.094 |
SLC7A14, SLC38A6 |
|
Cellular component |
|
GO:0005829 |
Cytosol |
25 |
1.881 |
0.002 |
CRTC3, CALCOCO2, SLC7A14, GPHN, SPATA7, GIGYF2, ZFYVE28, DOCK10, POGZ, ABL1, DENND5A, CERT1, HERC6, PRKCH, PLEKHA5, YLPM1, MED27, PLEKHA7, DAAM1, MTHFD1, DPYD, SERGEF, SLC28A1, SKAP1, RBMS3 |
|
GO:0005654 |
Nucleoplasm |
20 |
2.037 |
0.003 |
RNASEH2B, CRTC3, SLC7A14, PLEKHA5, RSF1, EFCAB6, PSEN1, MED27, PLEKHA7, SPATA7, DOCK10, MYO1C, CUX1, POGZ, DPF3, MAML3, SERGEF, CERT1, SKAP1, HERC6 |
|
GO:0015629 |
Actin cytoskeleton |
5 |
6.402 |
0.008 |
MACF1, ABLIM1, MYO1C, SWAP70, ABL1 |
|
GO:0048471 |
Perinuclear region of cytoplasm |
7 |
3.399 |
0.016 |
PPP1R16B, CALCOCO2, ABL1, PSEN1, SYNJ2BP, CERT1, FGFR3 |
|
GO:0005737 |
Cytoplasm |
27 |
1.547 |
0.017 |
MACF1, CRTC3, NCAPG, FMN1, IQGAP2, GPHN, FARS2, ABLIM1, DACH1, RELN, GMDS, RSRC1, RNF150, TNS4, GABBR2, PRKCH, SWAP70, KLHL1, GAB2, CDYL, SERPINB6, PSMA3, MYO1C, DPYD, PPP1R12B, SKAP1, GPHB5 |
|
GO:0016607 |
Nuclear speck |
6 |
3.853 |
0.019 |
PPP1R16B, RSRC1, YLPM1, MAML3, CDYL, SLC28A1 |
|
GO:0042995 |
Cell projection |
3 |
6.031 |
0.087 |
PPP1R16B, MACF1, STON1 |
Download Excel Table
Table 6.
The results of KEGG pathway analysis of candidate genes associated with total collagen contents trait from GWAS analysis
|
KEGG ID |
Description |
#Genes |
Fold enrichment |
p-value |
Gene name |
|
bta04810 |
regulation of actin cytoskeleton |
5 |
5.610 |
0.011 |
DIAPH3, IQGAP2, PPP1R12B, FGFR3, MYLK4 |
Download Excel Table
DISCUSSION
Meat tenderness of beef is one of the most important factors affecting meat quality evaluation of consumers [27]. The production of beef in consideration of a recent consumption pattern that favors tender meat with low fat is required. For this, it is necessary to produce tender meat with a short-term of breeding. This will make it possible to reduce production costs and protect the environment. We studied collagen contents that contribute to meat tenderness and texture for the production of tender meat. In particular, since collagen is greatly affected by genetics [28], it is thought to be suitable for use as a selection index to soft Hanwoo meat production through genomic analysis. Therefore, we conducted this study to detect significant SNPs and candidate genes associated with collagen contents trait using GWAS.
We identified a total of 73 significant SNPs and 108 candidate genes associated with total collagen contents trait. We identified TMEM135 and ME3 genes in which the most significant SNPs associated with collagen contents trait were located. TMEM135 gene was essential for collagen production and secretion in human cells [29]. In mouse hearts, forced overexpression TMEM135 can lead to collagen accumulation [30]. ME3 gene has beem found to be related to COL14A1, which plays a role in cross-linking collagen 1 and developing fibrous structure [31]. These collagen cross-links are regulated by myofibrillar protein and diverse gene expression related to muscle development. They determine meat tenderness [32]. Since we confirmed that these genes were related to collagen, we predicted that variants in these genes might affect meat tenderness of Hanwoo. We then predicted biological functions and pathways associated with candidate genes to find out how these genes were related to the collagen contents trait. In biological process of GO, we found that CRTC3 (Cyclic adenosine monophosphate [cAMP] -regulated transcriptional coactivator 3) and RELN genes were involved in positive regulation of CREB transcription factor activity (GO:0032793). CRTC3 is a coactivator of cAMP response element binding protein (CREB) that mediates the function of protein kinase A (PKA) signaling pathway. It is involved in various biological processes including lipid and energy metabolism. In porcine, CRTC3 expression is related to fat deposition in vivo. Furthermore, CRTC3 overexpression can increase lipid accumulation and the expression of mature adipocyte-related genes in cultured porcine subcutaneous adipocytes [33]. Lipid accumulation affects meat production and meat quality such as tenderness, juiciness, and flavor [34]. The deposition of subcutaneous and visceral fat directly influences backfat thickness and growth efficiency, while intramuscular fat (IMF) content directly affects meat quality including the flavor, juiciness, tenderness, and fatty acid (FA) composition [35]. In cellular component of GO and KEGG pathway, we found that candidate genes were involved in actin cytoskeleton (GO:0015629) and regulation of actin cytoskeleton (bta04810). A previous study has revealed that actin cytoskeleton-related cell junction is associated with lipid metabolism to influence the deposition of IMF. IMF is one important factor that can influence meat quality. A certain amount of IMF can enhance meat quality traits such as the flavor, juiciness, water holding capacity, and tenderness [36]. MYO1C (Myosin IC) and MYLK4 (Myosin Light Chain Kinase Family Member 4) genes belonging to these biological functions and pathways wererelated to meat tenderness. MYO1C has specialized functions in certain cell types such as muscles. Some researchers have linked myosins to meat tenderness [37]. MYLK4 regulates yak muscle contraction via phosphorylating myosin light chain molecules. Such phosphorylation can positively impact meat tenderness [38].
Variants and genes identified in this study are expected to provide important information for genomic selection of phenotypes to improve meat quality of Hanwoo. Furthermore, they provide a basis for further studies on consumption traits.