SHORT COMMUNICATION
Complete genome sequence of functional probiotic candidate Lactobacillus amylovorus CACC736
Soyeon Park
1
, Jung-Ae Kim
1,2
, Hyun-Jun Jang
1
, Dae-Hyuk Kim
1,3
, Yangseon Kim
1,*
1Department of Research and Development, Center for Industrializaton of Agricultural and Livestock Microorganisms, Jeongeup 56212, Korea
2Department of Bioactive Material Science, Jeonbuk National University, Jeonju 54896, Korea
3Department of Molecular Biology, Department of Bioactive Material Science, Institute for Molecular Biology and Genetics, Jeonbuk National University, Jeonju 54896, Korea
*Corresponding author: Yangseon Kim, Department of Research and Development, Center for Industrialization of Agricultural and Livestock Microorganisms, Jeongeup 56212, Korea. Tel: +82-63-536-6712, E-mail:
yangseon@cialm.or.kr
© Copyright 2023 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Sep 28, 2022; Revised: Oct 10, 2022; Accepted: Oct 13, 2022
Published Online: Mar 31, 2023
Abstract
Lactobacillus amylovorus CACC736 was originated from swine feces in Korea. The complete genome sequences of the strain contained one circular chromosome (2,057,809 base pair [bp]) with 38.2% guanine-cytosine (GC) content and two circular plasmids, namely, pCACC736-1 and pCACC736-2. The predicted protein-coding genes, which are encoding the clustered regularly interspaced short palindromic repeats (CRISPR)-associated proteins, biosynthesis of bacteriocin (helveticin J), and the related proteins of the bile, acid tolerance. Notably, the genes related to vitamin B-group biosynthesis (riboflavin and cobalamin) were also found in L. amylovorus CACC736. Collectively, the complete genome sequence of the L. amylovorus CACC736 will aid in the development of functional probiotics in the animal industry.
Keywords: Lactobacillus amylovorus; Swine; Probiotics; Whole-genome sequencing
Lactobacillus spp. are non-pathogenic microorganisms that provide beneficial effects to the host [1–3]. Lactobacillusamylovorus has been studied as a paraprobiotic (non-viable cells or cell fractions) with the ability to change body adiposity [1]. Additionally, it has been reported that L. amylovorus has probiotic properties such as antiviral and antimicrobial activities through the regulation of the gut microflora [2,3]. In this study, the genomes of L. amylovorus CACC736 are functionally annotated.
L.amylovorus strain CACC736 (KACC22146) was isolated from swine feces in Korea. This strain was inoculated in de Man, Rogosa, and Sharpe (MRS) medium (Difco, Franklin Lakes, NJ, USA) and cultivated at 37°C for 24 h. Genomic DNA (gDNA) of the strain was extracted using the DNeasy UltraClean microbial kit (Qiagen, Hilden, Germany). The complete genome sequence of L.amylovorus strain CACC736 was obtained with single-molecule real-time sequencing technology (SMRT) on the platform of PacBio Sequel (Pacific Biosciences, Menlo Park, CA, USA) at CJ Bioscience, Inc (Seoul, Korea). These gene sequences were performed by de novo genome assembly using the PacBio SMRT Analysis (version 2.3.0, Pacific Biosciences) [4]. All genes were classified by different functional groups using EggNOG 4.5 (http://eggnog5.embl.de). Additionally, functional annotation of the coding sequences (CDSs) was performed by the UBLAST program including the databases of the Swiss-Prot and Kyoto Encyclopedia of Genes and Genomes (KEGG) [5]. Predictions for clustered regularly interspaced short palindromic repeats (CRISPR) were used by CRISPR finder (https://crispr.i2bc.paris-saclay.fr) [6].
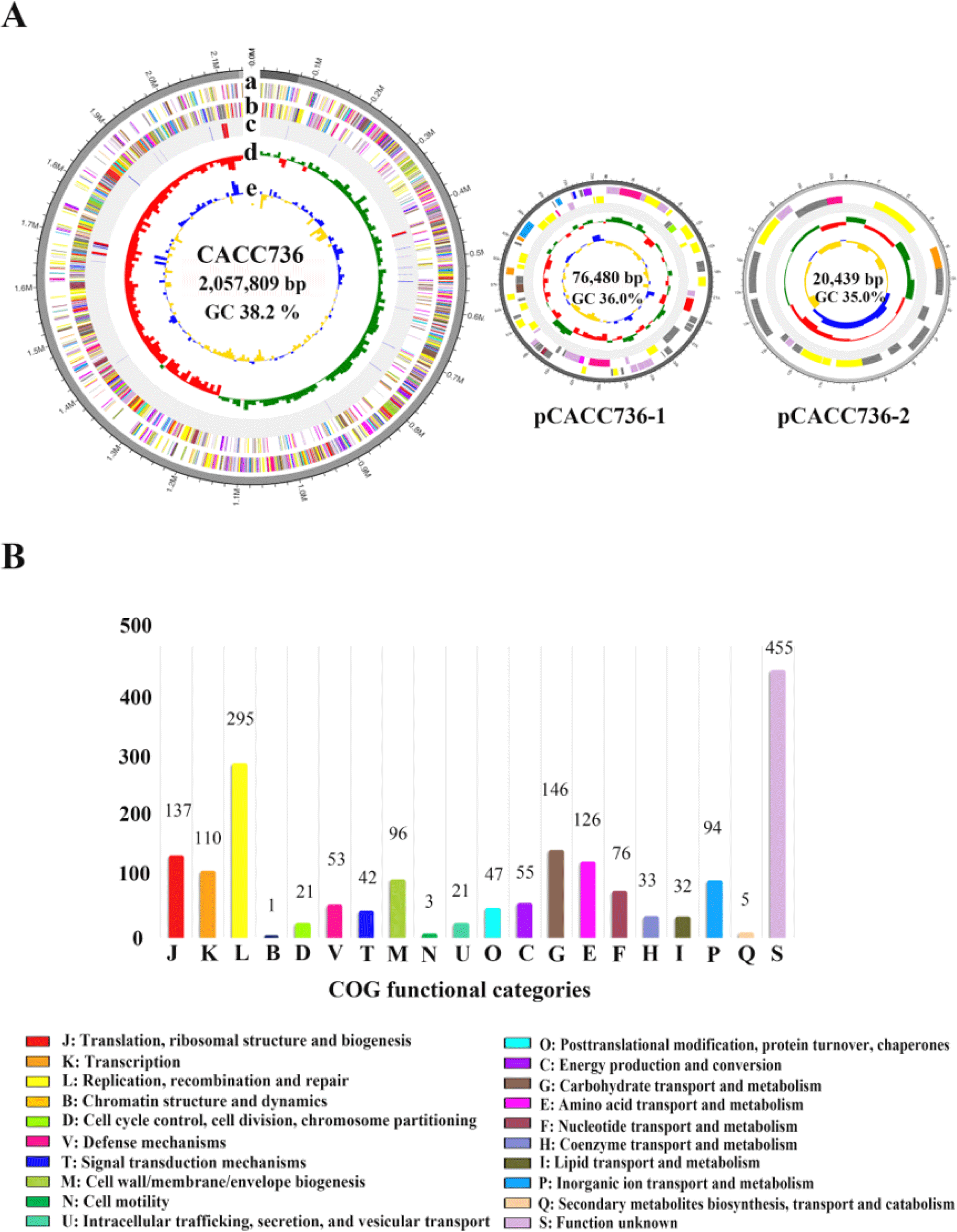
The L.amylovorus CACC736 composed of one circular chromosome (2,057,809 base pair [bp], 38.2% guanine-cytosine [GC] content) along with two plasmids designated as pCACC736-1 (76,480 bp, 36.0% GC content) and pCACC736-2 (20,439 bp, 35.0% GC content) (Table 1 and Fig. 1A). Moreover, the complete genome comprised 2,080 protein- CDSs and 80 non-coding genes (15 rRNA and 65 tRNA genes). A total of 1,848 proteins (88.8%) were classified on a functional categorization by the database of Clusters of Orthologous Groups (COGs) categories (Fig. 1B). The most abundant COGs categories, excluding an ‘unknown function [S]’, were ‘replication, recombination and repair [L]’ (295 genes; 16.0%), ‘carbohydrate transport and metabolism [G]’ (146 genes; 7.9%), ‘translation, ribosomal structure and biogenesis [J]’ (137 genes; 7.4%), and ‘amino acid transport and metabolism [E]’ (126 genes; 6.8%). The genome of the L. amylovorus CACC736 encoded five CRISPR genes/proteins (Cas1, Cas2, Cas3, Cas4, and Cas6) for antiviral-relative mechanisms [7], one bacteriocin class III (helveticin J) for an inhibitory effect against common pathogenic organisms [8], and two potential genes of antimicrobial activity (lysM and qac). In addition, the L.amylovorus CACC736 was confirmed to have genes associated with common probiotic properties, such as lactate synthesis (ldh, L-lactate dehydrogenase), bile salt hydrolases (BSH; cbh) and acid tolerance (atpD, atpH, and grpE) (Table 2). Notably, we revealed the presence of genes involved in vitamin B2 and B12 biosynthesis, including riboflavin (ribB, ribD, ribE, and ribT) and cobalamin (cobC) (Table 2) [9,10]. Taken together, our findings on the complete genome of L.amylovorus CACC736 will provide a scientific improvement for the development of functional probiotics.
Table 1.
General features of Lactobacillus amylovorus CACC736 genome
|
Properties |
Chromosome |
Plasmids |
|
CACC736 |
pCACC736-1 |
pCACC736-2 |
|
BioProject |
PRJNA881772 |
− |
− |
|
BioSample |
SAMN30915630 |
− |
− |
|
Accession No. |
CP104879 |
CP104880 |
CP104881 |
|
Genome size (bp) |
2,057,809 |
76,480 |
20,439 |
|
GC content (%) |
38.2 |
36.0 |
35.0 |
|
No. of CDSs |
1,989 |
71 |
20 |
|
No. of CRISPR regions |
5 |
− |
− |
|
rRNA genes |
15 |
− |
− |
|
tRNA genes |
65 |
− |
− |
Download Excel Table
Table 2.
Predicted CDSs involved in probiotic potency in Lactobacillus amylovorus CACC736
|
Predicted function |
L. amylovorus CACC736 |
|
Predicted genes |
Start position |
End position |
Length (bp) |
|
CRISPR/cas |
|
Endonuclease |
Cas1 |
c1,507,114 |
c1,508,103 |
990 |
|
Endonuclease |
Cas2 |
c1,506,827 |
c1,507,108 |
282 |
|
Endonuclease/helicase |
Cas3 |
c1,508,620 |
c1,511,049 |
2,430 |
|
Exonuclease |
Cas4 |
c1,508,113 |
c1,508,604 |
492 |
|
Endoribonuclease |
Cas6 |
c1,515,924 |
c1,516,679 |
756 |
|
Antimicrobial activity-related |
|
Lysin motif domain |
lysM
|
890,591 |
891,055 |
465 |
|
Quaternary ammonium compound-resistance |
qacC
|
1,012,642 |
1,012,962 |
321 |
|
Bacteriocin (Class III) |
helveticin J |
c1,995,360 |
c1,995,992 |
633 |
|
Lactate synthesis |
ldh
|
1,795,954 |
1,796,925 |
972 |
|
Bile salt hydrolases (BSH) |
cbh
|
1,052,335 |
1,053,357 |
1,023 |
|
Acid tolerance-related |
atpD
|
690,181 |
691,692 |
1,512 |
|
atpH
|
692,699 |
694,138 |
1,440 |
|
clpB
|
98,750 |
100,879 |
2,130 |
|
grpE
|
c1,235,180 |
c1,235,764 |
585 |
|
Protection or repair-related |
dnaJ
|
c1,232,063 |
c1,233,217 |
1,155 |
|
Vitamin B-groups synthesis |
|
Vitamin B2
|
ribB
|
c1,025,985 |
c1,027,160 |
1,176 |
|
ribD
|
c1,027,752 |
c1,028,810 |
1,059 |
|
ribE
|
c1,027,163 |
c1,027,759 |
597 |
|
ribT
|
887,197 |
887,550 |
354 |
|
Vitamin B12
|
cobC
|
c301,776 |
c302,426 |
651 |
Download Excel Table
Fig. 1.
Genome futures of Lactobacillus amylovorus CACC736.
(A) Circular genome mapping of Lactobacillus amylovorus CACC736. Circles from the outside to the center denote: (a) forward and (b) reverse strands (colored according to COGs function categories), (c) rRNA and tRNA, (d) GC skew, (e) GC content. (B) Functional classification of COGs. COGs, cluster of orthologous groups of proteins database; GC, guanine-cytosine.
Download Original Figure
NUCLEOTIDE SEQUENCE ACCESSION NUMBERS
The complete genome sequences of L.amylovorus strain CACC736 were deposited at the NCBI GenBank under the accession numbers CP104879 (chromosome) and CP104880-CP104881 (plasmids), respectively.