INTRODUCTION
Native pig breeds are very important for the conservation and sustainable improvement of valuable economic traits in the future [1]. Native pigs in Korea, especially Jeju native pigs (JNP), have been extensively studied for crossbreeding with commercial pig breeds such as Landrace and Duroc and for quantitative trait locus (QTL) and genome-wide association study (GWAS) for identifying genetic markers of economic traits. The coat color of JNP is black, and the feed efficiency and growth rate of JNP are low. However, JNP has excellent meat quality characteristics, such as a solid fat structure, red meat color (a*), and high intramuscular fat (IMF) content, compared with commercial pig breeds, including Landrace [2].
Collagen is a major substrate protein in connective tissues such as skin, tendon, bone, and blood vessels in animals [3,4]. Collagen is a lightweight protein that exists in epithelial cells and is widely distributed in multicellular animals such as invertebrates and vertebrates [5]. Collagen can form insoluble fibers with high tensile strength and has a triple hypochondrobar structure consisting of three identical polypeptide chains. Collagen is also the most abundant protein in vertebrates and makes up approximately 25% of the total protein in vertebrates [6]. Collagen comprises approximately 70% of the bones of organisms, approximately 50% of the cartilage connecting bones with bones in joints, approximately 70% of the dermis under the skin, and most vessels [7]. Collagen fibers exist in other forms between the perimysium and endomysium and are mainly composed of thick bundles of collagen in the perimysium and a fine network structure in the endomysium. Fibers depend on the type and area of collagen in the muscle, and collagen content’s quantitative and chemical composition changes over time [8]. Collagen is classified according to its composition. Type I is the most abundant and strongest type of collagen found in the human body. It consists of eosinophilic fibers that form tendons, ligaments, organs, and skin. Type I collagen helps form bones and can be found in the gastrointestinal tract. In addition, collagen plays a major role in healing wounds, giving skin elasticity, and maintaining tissues. Type II collagen is mainly found in connective tissues and plays a role in forming cartilage. The health of the joints depends on cartilage composed of Type II collagen, which helps prevent various arthritis symptoms. Type III collagen is the main component of the reticular fiber and the extracellular matrix that makes up organs and skin. Collagen is found mainly in the Type I form and plays a role in skin elasticity and hardness. In addition, collagen forms blood vessels and tissues in the heart, so a deficiency in Type III collagen increases the risk of vascular rupture and premature death. Type IV collagen plays an important role in forming the basal lamina found in endothelial cells that form tissues surrounding organs, muscles, and fats, which cushion and protect tissues in the space between the top and bottom layers of the skin. The basal plate is necessary for the function of various nerves and blood vessels, so it constitutes most of the digestive organs and respiratory surfaces in the body. Type V collagen is needed to make up the cell surface as well as female placental tissue and hair strands [9,10].
Collagen protein, a byproduct of meat production, is an important ingredient in food products such as casings. In addition, collagen is used in the production of cosmetics and is widely used as a healing aid following plastic surgery, bone reconstruction, and various dental and orthopedic surgeries. Collagen is used for cosmetic purposes to treat wrinkles and skin aging, as well as in vaccines and vitamins [11]. In addition, collagen has attracted attention from pharmaceutical and biomaterial-based packaging industries, as it can be used to encapsulate and form edible films [3]. Commercial collagen used for biomedical applications is extracted mainly from the skin of pigs and cattle. However, the outbreak of prion diseases, such as bovine spongiform encephalopathy, has resulted in anxiety among users of collagen derived from cattle. Interest in safer sources of collagen, such as pigs, has greatly increased [12].
Recently, we reported evidence that a 6-bp deletion (XM_013981330.2: g.−1805_−1810del) in 5´-regulatory region of myosin heavy chain 3 (MYH3) is the first functional sequence variant (FSV) for red meat color (a∗) and IMF in domestic pigs [2]. We firstly mapped a 488.1-kb critical region in porcine chromosome 12 that influences both a* and IMF by a combined linkage-linkage disequilibrium analysis in two independent F2 intercross between Korean native pigs (KNPs) and Western commercial breeds (i.e., Landrace and Duroc). In this critical region, only the MYH3 gene, encoding myosin heavy chain isoform 3, was found to be discriminatingly overexpressed in the skeletal muscle of KNPs than in that of Landrace pigs. Subsequently, the MYH3 gene was verified as a causal gene for the two traits using transgenic mice, and then detected the 6-bp deletion (XM_013981330.2: g.−1805_−1810del) variant in the 5´-promoter region of the MYH3 gene for which Q allele carriers showed significantly higher values of a* and IMF than q allele carriers. Additionally, we demonstrated that this 6-bp deletion variant could abrogate the binding of the regulatory myogenic regulatory factors (MRFs, i.e., MYF5, MYOD, MYOG and MRF4) and act as a significantly weaker repressor, leading to increased expression of the MYH3 gene in the skeletal muscle. Biological mechanism regarding myosin heavy chain and collagen has not well been known. Recently, Coelho et al. reported that interaction of discoidin domain receptor 1 (DDR1) with myosin motors can contribute to collagen remodeling [13]. However, biological relationship between the FSV of MYH3 possessing the domain of myosin motors and collagen content needs to be further investigated. Therefore, the aim of this study was to examine the association between FSVs located in the 5´-upstreme region of the MYH3 gene and collagen content in crossbred JNP and Landrace pigs.
MATERIALS AND METHODS
All experimental procedures were conducted according to national and institutional guidelines and approved by the Ethical Committee of the National Institute of Animal Science, Republic of Korea (Approval number: 2017-241).
A total of 187 animals of the F4 resource population (103 male and 84 female) were established by crossing F3 animals derived from intercrossing JNP and Landrace pigs. Animals were raised at the experimental farm of the National Institute of Animal Science, Jeju, Korea. They were fed ad libitum, and males were not castrated. All F4 experimental animals were slaughtered in the same commercial slaughterhouse (Jeju Livestock cooperatives, Jeju, Korea). The range of slaughter age of the pigs used for collagen content analysis was 180-200 days. The Musculus longissimus dorsi, Musculus semimembranosus, Musculus triceps brachii and M. biceps femoris from each carcass were used for the analysis of collagen content [14].
Genomic DNA was isolated from blood and muscle using the sucrose-proteinase K method and used as a template for polymerase chain reaction (PCR) [15]. The absorbance of the separated DNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and then genomic DNA with an A260/A280 ratio of 1.8 or more was used as a template for PCR.
The collagen content was analyzed according to the method described by Choi et al. [16]. After 4 g of ground meat sample was placed into a triangular flask, 30 mL of a sulfuric acid solution was added. The sample was covered with a triangular flask in a dry oven for 16 hours before measurement. The hydrolyzable material was put in a 500 mL flask, diluted with distilled water, and filtered in a 100 mL triangular flask. 5 mL of the filtered solution was taken and diluted in 100 mL. Then, 2 mL of the final diluent was put in a 10 mL test tube, and 1 mL of the oxidizing solution was added. In the blank, 2 mL of distilled water and 1 mL of an oxidation solution were added instead of the diluent. samples were incubated at room temperature for 20 minutes, dissolved in 4-dimethyl-aminobenzaldehyde (60% w/w) in a color reagent (35 mL perchloric acid), mixed with 1 mL of 2-propanol 65 mL), covered with a cap, and immediately heated in a constant-temperature water bath (60°C) for approximately 15 minutes. The amount of hydroxyproline was measured from the standard curve, and the collagen content (%) was calculated by multiplying the hydroxyproline content by a constant of 8.
A set of primers was designed for the detection of the FSVs in the MYH3 gene using available sequence information (XM_013981330.2). PCR was performed using a Maxime PCR Premix kit, and reactions included 20 µL of reaction mixture including 100 ng of DNA, 0.5 nmol of MYH3 promoter-specific primers (MYH3_1_F, 5’-TGGTCTTTCCTAATTGGTGACAT-3’, MYH3_1_R, 5’-AGTTTTGAGCAAGGCTTTTGTT-3’) and distilled water (iNtRON, Seongnam, Korea). PCR conditions were as follows: initial heating was at 95°C for 5 min, followed by 35 cycles of 30 s for denaturation at 94°C, 30 s for annealing at 65°C, and 30 s for an extension at 72°C, followed by a final extension at 72°C for 10 min in a Nexus PCR machine (Eppendorf, Hamburg, Germany). And the amplicons were digested with the restriction enzyme, HpyCH4IV (NEB, Ipswich, MA, USA). The PCR products were separated on 2.5% agarose gels (Lonza, Basel, Switzerland) and visualized by UV illumination with a BioFACT 100 bp plus DNA ladder marker (BioFACT, Daejeon, Korea) [2].
The allele frequencies and heterozygosity (he), polymorphic information contents (PIC), and χ2 values (p-values) for MYH3 FSVs were calculated using the CERVUS 3.0.3 program [17].
Putative outliers were deleted based on the ascertainment of normality using a Ryan-Joiner (RJ) evaluation in the MINITAB program (Minitab, State College, PA, USA). An RJ score ≥ 0.99 was used to ascertain normality. The following general linear model (GLM) was fitted to the phenotype data to estimate the effect of the FSVs of MYH3 on collagen content using MINITAB:
where, Yiik is the observed phenotype, μ is the mean value, genotypei represents the FSVs in the MYH3 gene (Q/Q, Q/q, and q/q), X is the carcass weight as a covariate, b1 is the regression coefficient and eijk is the random residual. We used Tukey’s multiple comparison method to separate means and set the significance level at p < 0.05. The percent phenotypic variance of a trait explained by the MYH3 FSV was computed by [(VARreduced−VARfull)/VARreduced] × 100, where VARreduced and VARfull are the variances of residuals of a GLM without and with genotypes of the MYH3 FSV in the models.
RESULTS AND DISCUSSION
The FSVs in the MYH3 gene were genotyped using PCR-restriction fragment length polymorphism (RFLP) in the Landrace and JNP crossbred population. Determination of the genotype of the FSV of the MYH3 gene was conducted by PCR amplification and subsequent HpyCH4IV digestion. The q/q genotype represents the MYH3 homozygous genotype originating from Landrace and Duroc pigs; the Q/Q genotype represents the MYH3 homozygous genotype originating from JNPs. The results indicated that both the Q and q alleles were segregating, and all three genotypes (QQ, Qq, and qq) were identified for the FSVs of MYH3 (Table 1 and 2, Fig.1; Q and q are the Landrace and JNP variants, respectively).
| FSVs of MYH3 genotype frequency | χ 2 1) | Allele frequency | Diversity parameter | |||||
|---|---|---|---|---|---|---|---|---|
| QQ (67) | Qq (103) | qq (17) | Q | q | Ho | He | PIC2) | |
| 0.358 | 0.551 | 0.091 | 5.7260.01 | 0.634 | 0.366 | 0.551 | 0.466 | 0.357 |
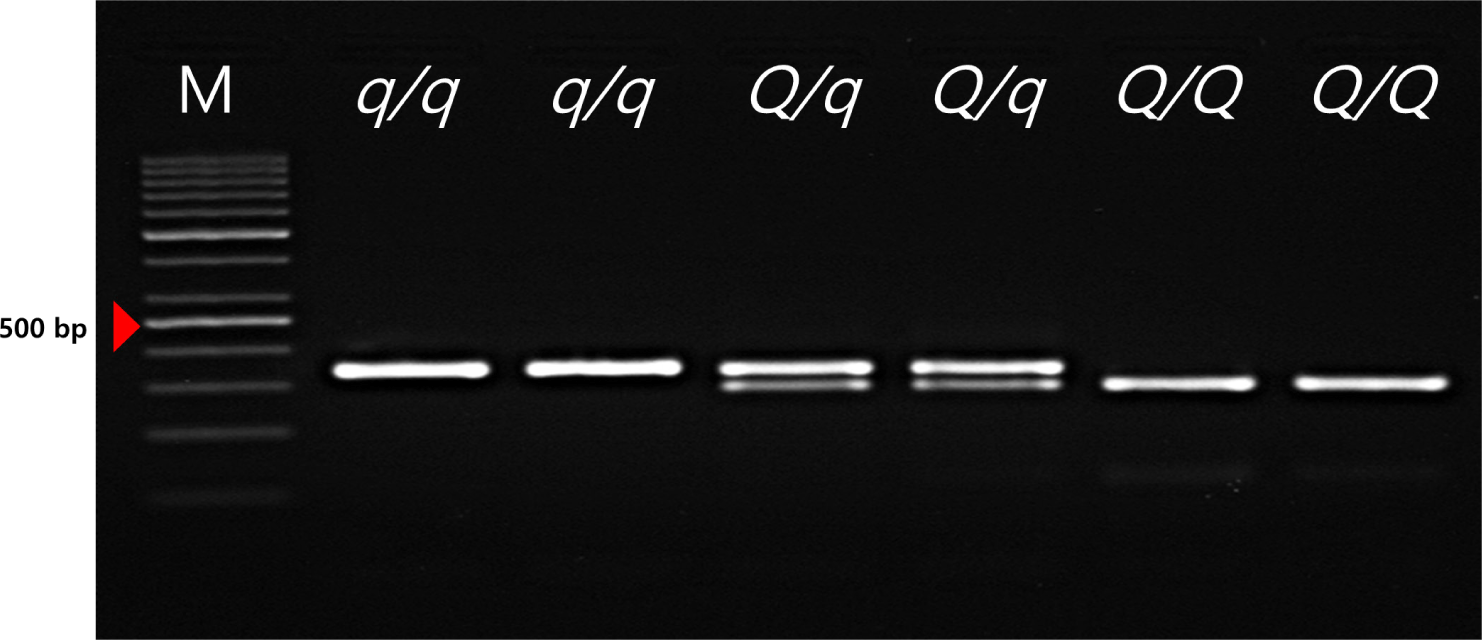
These FSVs in the MYH3 gene had allele frequencies of 0.634 and 0.366 for the Q and q alleles, respectively. This indicates a higher frequency of the Q allele than the q allele because the favorable Q allele was selected for in this population. The FSVs in the MYH3 genotype had frequencies of 0.358, 0.551, and 0.091 for QQ, Qq, and qq, respectively, indicating more heterozygous animals than homozygous animals were observed in this population. However, the chi-square test results revealed that the FSVs were not in Hardy–Weinberg equilibrium (p < 0.05), indicating that artificial selection was applied to the QQ genotype animals used in this study. These results confirm our expectations that the selection pressure was applied to obtain more favorable alleles in the population. In addition, the PIC value was found to be 0.357, suggesting that this population showed intermediate polymorphism for these FSVs (Table 2). In a previous study to identify this causal variant, the Q allele is associated with higher redness value (a*), increased IMF contents, higher Type1 muscle fiber area and increased myoglobin content compared to the q allele in the Landrace × JNP crossbred pig population [2,18]. Therefore, we can improve the meat quality of JNP × Landrace crossbred population via the marker-assisted selection (MAS) of the FSV of MYH3.
The MYH3 gene encodes a portion of myosin, a contractile protein, which is especially fundamental to the proper functioning the Sarboomer of striated/skeletal muscle. MYH3 is also recognized as an embryonic myosin heavy chain because it was mainly overexpressed in early mammalian development [19].
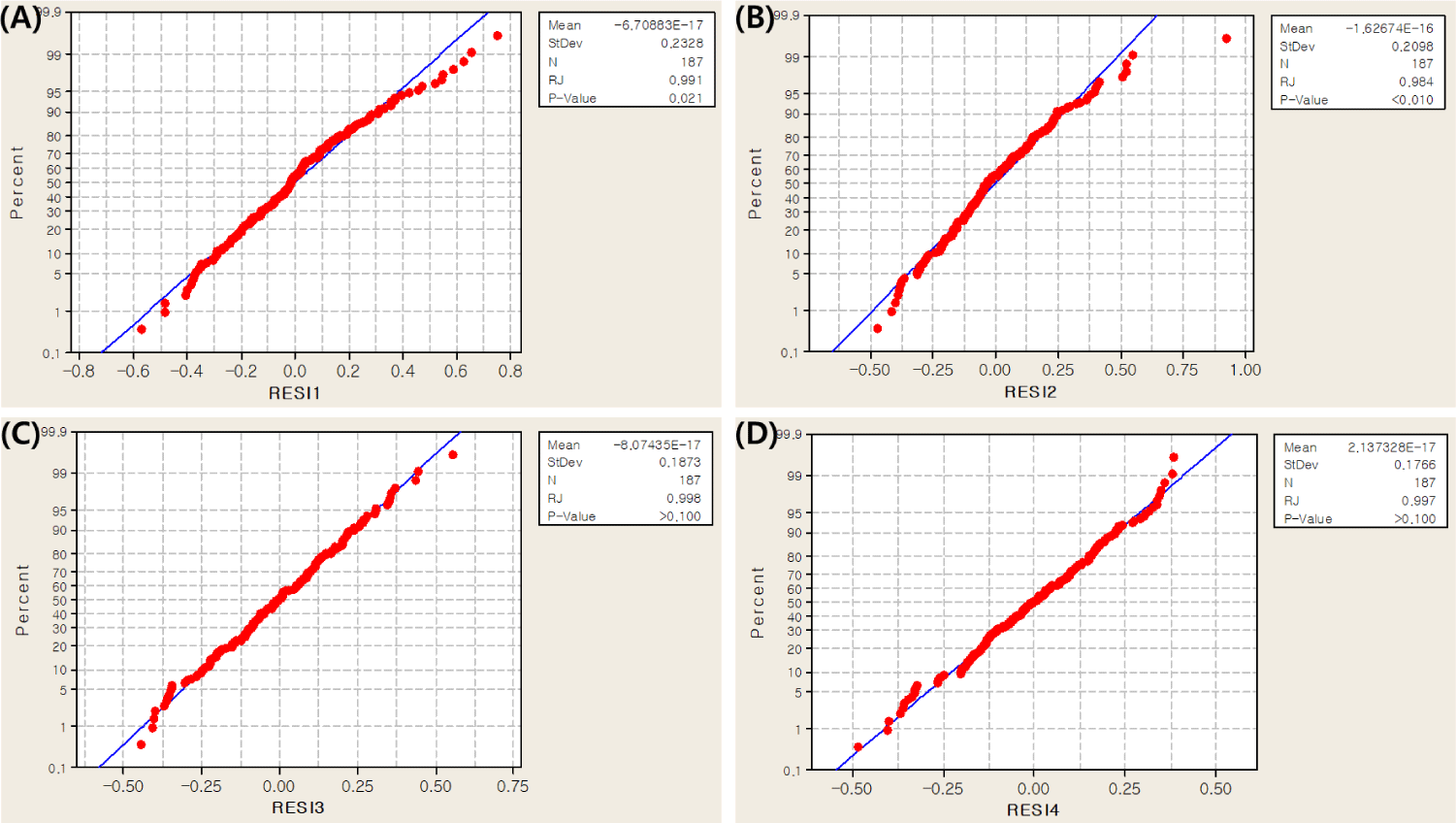
In this study, collagen content was measured in four muscles (M. longissimus dorsi, M. semimembranosus, M. triceps brachii and M. biceps femoris) in the Landrace and JNP crossbred population. Before the association test, a normality test for collagen content was performed using the RJ method [20]. The results showed an RJ score of 0.990 or higher, indicating that the collagen content data followed a normal distribution and could be further analyzed (Fig. 2).
Through analysis of the association between collagen content and FSVs of the MYH3 gene, we found that individuals with the QQ genotype had higher collagen content than those with the qq genotype in M. longissimus dorsi, M. semimembranosus, M. triceps brachii and M. biceps femoris muscles (Table 3, p < 0.05, p = 2.00×10−16, p = 6.66×10−16 and p = 3.31×10−5, respectively).
Pork collagen has been used since the beginning of artificial skin research, primarily to assist in the treatment of burns and trauma. When a wound occurs on the skin, a temporary protective film such as a silicon dressing is attached until the damaged skin recovers. This prevents water leakage from the body, absorbs exudate (liquid coming out of the blood when there is inflammation), and prevents invasion and infection by bacteria from the outside. Except for the application of a gel, the dressing is a porous membrane made using a polyurethane membrane or chitin, a freeze-dried product of pig leather, or the like [12]. However, it is applied in limited situations and is expensive. When we computed the percent variance of the MYH3 FSV as the effect size, we found that the MYH3 FSV explained up to 39.7% of phenotypic variance (Table 3). This amount of effect size of MYH3 FSV can be regarded as a good indication that this FSV could be used in the MAS for improving collagen content in this JNP × Landrace crossbred population. With the fixation of QQ animals, the collagen content in pork can be improved, meaning that consumers can have better quality pork and that the pork industry can benefit from increasing the value of nonpreferred pork collagen, which can be used for medical purposes. However, it is necessary to conduct additional experiments in other independent populations to confirm the effect size of MYH3 FSV before executing the MAS for collagen content. Selection for pigs with more collagen may allow this research to be applied in the biomedical area and provide profits to farmers.
