INTRODUCTION
Ruminants are an important source of most meat and dairy products consumed by people worldwide [1–5]. Unlike monogastric animals, the ruminant animals consumed and fermented the feeds containing plant materials in the rumen, an anaerobic chamber in which various microorganisms are digested and absorbed [6–8]. The rumen fluids in the anaerobic fermentation chamber mainly contain a wide range of anaerobic bacteria, protozoa, fungi, methanogenic archaea, and phages [9]. During fiber degradation metabolism, various rumen microorganisms release a variety of metabolites and vesicles through anaerobic fermentation [10,11].
Extracellular vesicles (EVs) are nanosized vesicles released by all cells, including bacteria [12,13]. EVs are lipid bilayer-enclosed structures containing proteins, lipids, and nucleic acids. And they belong to a large family of exosomes (30–200 nm), microvesicles (100–350 nm), and apoptotic blebs (500–1000 nm) [14,15]. According to recent studies, EVs mediate cell and organ interactions because they contain DNA, RNA, proteins, and lipids and have the potential as biomarkers and targeted therapeutic mediators [16]. Recent studies reported that all kinds of bacteria could produce EVs. The existence of bacteria-derived EVs in body fluids such as milk, blood, sweat, urine, or feces suggests that these molecules have an impact on host cells and organs by directly activating host receptors, conveying different bioactive chemicals or integrating EVs into host cells [17–20]. In particular, in the livestock industry, EVs are widely used as biomarkers for monitoring bovine mastitis, stress conditions, and different diseases in livestock [21–23]. The Caenorhabditis elegans surrogate model has been widely used to understand conserved mechanisms in host-EV interactions. C. elegans is a multicellular animal model that has the potential to close the gap between in vitro and in vivo approaches by genetic tractability, simplicity of culture, and comparatively short lifespan [24,25]. Therefore, we used the genetically tractable model host C. elegans to investigate the effect of EVs on the host.
To date, numerous studies on the function of the various microorganisms in the rumen and their interactions have been conducted, but EVs released by rumen microorganisms and their interactions with the host are still poorly understood. Consequently, this study aimed to identify rumen EVs (REVs) and investigate REVs-host interactions using a C. elegans animal model.
MATERIALS AND METHODS
The isolation of REVs was performed using the standard differential ultracentrifugation method described in previous studies with some modifications [26]. In brief, bovine rumen was centrifuged at 5,000×g at 4°C for 15 min to remove large particles. The supernatant was centrifuged at 7,000×g at 4°C for 30 min to remove any remaining debris and cells. Then, the supernatant was passed through a 0.45 μm filter and a 0.22 μm filter to remove residual cell debris. The clear supernatant was centrifuged at 150,000×g at 4°C for 60 min using an ultracentrifuge. And we collected pelletized EVs, resuspended them in phosphate-buffered saline (PBS), and kept them at −80°C. For the preparation of sonication REVs samples (SREVs) as negative control, EVs were sonicated to 50 w (1 min for 8 cycles) using an ultrasonic homogenizer on ice before use for the experiment. The experimental protocol for this study was reviewed and approved by the Institutional Animal Care and Use Committee of Jeonju University (approval jjIACUCU-2022-0409-A1).
To characterize EVs, transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) were performed. TEM investigations were performed according to a previously described method [27,28]. A 5 μL EV sample was placed onto a carbon-coated grid and allowed to settle for 60 sec. The samples were negatively stained with 2% uranyl acetate. The samples were visualized by TEM at NICEM (The National Instrumentation Center for Environmental Management, Seoul National University, Korea). EV size distribution and number were assessed by NTA using a NanoSight NS300 (Malvern). The data were analyzed using NTA software version 3.4 Analytical.
C. elegans was maintained on nematode growth medium (NGM) agar plates supplemented with Escherichia coli OP50 strain at 20°C by using a standard protocol [29]. For the lifespan experiment, we used the CF512 fer-15(b26)II;fem-1(hc17) IV (fer-15;fem-1 worms) strain. In the lifespan assay, the L4 young adult stage worms were exposed to OP50 with EVs (approximately 1.0×1010 beads), and the number of live and dead worms was recorded every day. For the killing assays, L4 young adult stage worms were preconditioned with OP50 and EVs (1×1010) for 24 h and then transferred to Escherichia coli O157:H7 EDL933 and Staphylococcus aureus RN6390 as pathogenic bacteria. Then, living worms were counted every day.
For C. elegans transcriptome analysis, L4 young adult worms were exposed to EVs for 24 h. After 24 h, worms were harvested in M9 buffer and TRIzol for RNA isolation. After homogenization, RNA was extracted using a Qiagen RNeasy kit according to the manufacturer’s instructions. Total RNA concentration was measured using Quant-IT RiboGreen (Invitrogen, Waltham, MA, USA). To determine the quality of samples total RNA, samples are run on the TapeStation RNA screentape (Agilent Technologies, Santa Clara, CA, USA). For the construction of RNA libraries, we using high-quality RNA preparations (RIN > 7.0). Each sample’s library was individually prepared using the Illumina TruSeq Stranded Total RNA Library Prep Gold Kit (Illumina, San Diego, CA, USA). First, removing the rRNA from the total RNA. The remaining mRNA is broken up to little fragments using divalent cations at a high temperature. With the use of random primers and SuperScript II reverse transcriptase from Invitrogen, the cleaved RNA fragments are converted into first-strand cDNA. Next, Second-strand cDNA synthesis by utilizing DNA Polymerase I, RNase H, and dUTP. These cDNA fragments are next subjected to an end repair procedure, a single “A” base addition, and adapter ligation. The final cDNA library is made by purifying and enriching the results with polymerase chain reaction (PCR). Then, using KAPA Library Quantification kits for quantified libraries accroding to the quantitative PCR (qPCR) Quantification Protocol Guide (KAPA BIOSYSTEMS) and validated using TapeStation D1000 ScreenTape (Agilent Technologies). After the indexing of libraries, paired-end (2×100 bp) sequencing was conducted on a Illumina NovaSeq (Illumina).
The C. elegans lifespan and killing assay were analyzed according to the Kaplan–Meier (KM) method. A log-rank test between survival curves was performed using STATA software to examine the significance of differences (USA). A significant difference was defined as one that showed a significance (p-value 0.05) through all replicates.
RESULTS
In the TEM image, membrane vesicles with various sizes were observed (Fig. 1A). No EV was observed in samples treated with sonication (Fig. 1A). NTA was used to examine the size, distribution, and concentration of EVs. EVs in the rumen have an average size of 157.4 nm, which is considered to include microvesicles, microparticles, ectosomes and exosomes (Fig. 1B).
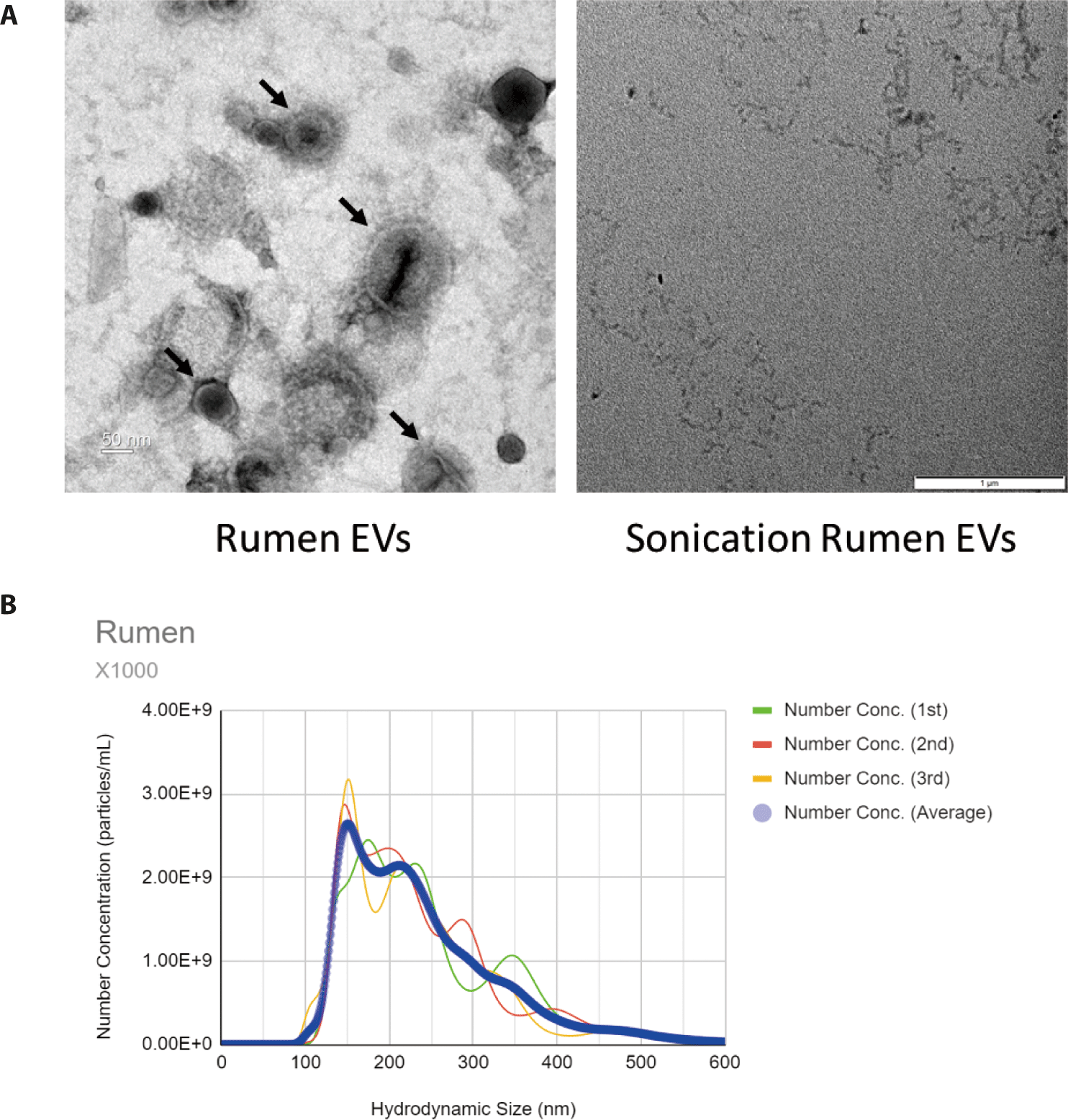
To evaluate the interactions between REVs and the host, we used the C. elegans animal model as an in vivo model. First, we investigated the effect of REVs on the lifespan of C. elegans. The lifespan of nematodes was not affected by exposure to REVs compared to the control E. coli OP50 alone (Fig. 2A). Next, we investigated whether REVs could increase the C. elegans defense response to pathogenic bacteria. We exposed C. elegans to E. coli OP50 with REVs and SREVs for 24 h, transferred the worms to E. coli O157:H7- and S. aureus-pathogenic bacteria-treated plates, and counted the viability every day. Contrary to the lifespan results, REVs significantly increased the worm’s lifespan when C. elegans was exposed to pathogenic bacteria (Fig. 2B). However, treatment with SREVs did not increase the lifespan of the worms. These findings imply that REVs enhance the survival of C. elegans from exposure to pathogenic bacteria.
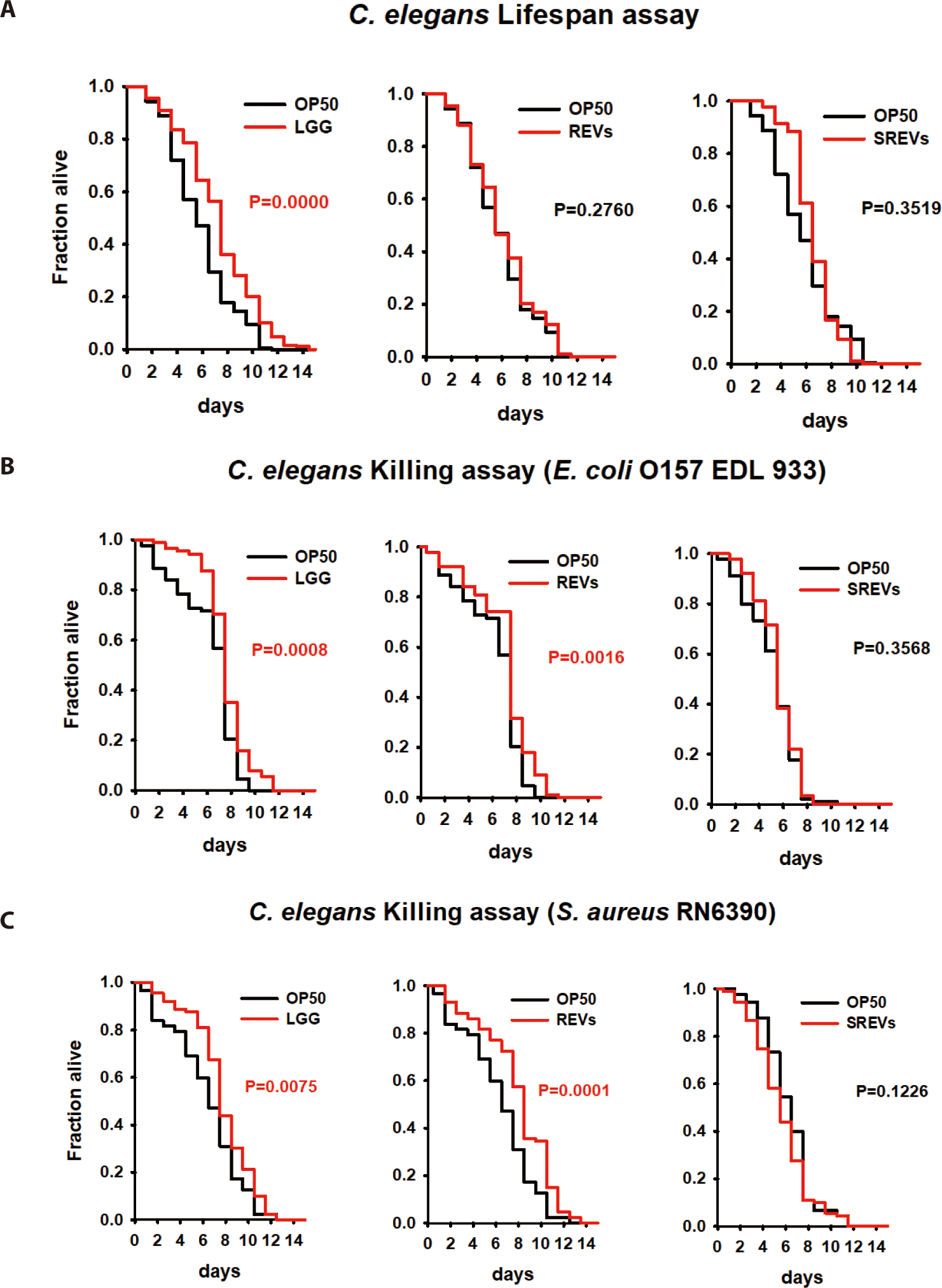
Since REVs induced C. elegans protection from pathogenic bacteria, we performed an RNA sequencing assay to investigate the genes regulated by exposed REVs. RNA from worms exposed to REVs (treatment group) and worms exposed to SREVs (control) were sequenced. Here, we focused on investigating transcript level changes on the previously unreported effects of REV’s treatment. Gene expression levels modulated by REVs and SREVs treatments were shown (Fig. 3A). REVs treatment in worms could significantly increase 433 genes and decrease 643 genes compared to the SREVs-treated group. Notably, REVs upregulated genes related to an olfactory receptor, chitinase activity, and peptide receptor activity, and downregulated genes related to double-stranded DNA binding, adenylate cyclase activity, and phosphoprotein phosphatase activity (Fig. 3B).
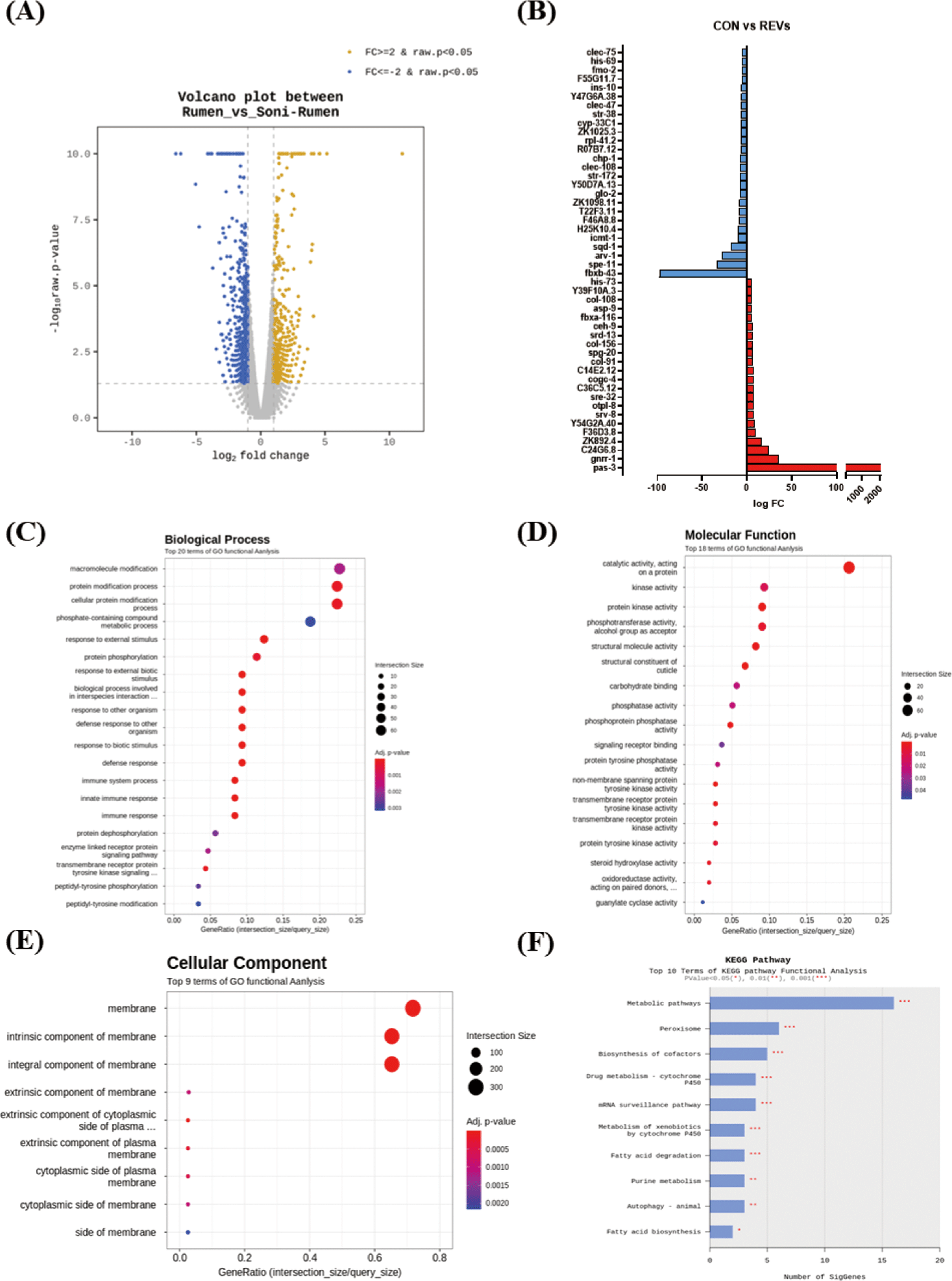
For an in-depth understanding, the results of enrichment analysis through the Gene Ontology (GO) Database for significant genes are shown as a dot plot. The REVs treatment in C. elegans could significantly regulate 69 GO terms. The 42 terms were for biological process (BP) (Fig. 3C), 18 for molecular function (MF) (Fig. 3D), and 9 for cellular component (CC) (Fig. 3E). The main GO terms in BP were GO:0044419 (biological process involved in interspecies interaction between organisms), GO:0098542 (defense response to other organism), and GO:0006955 (immune response). The most significantly enriched MFs were GO:0042302 (structural constituent of cuticle), GO:0004715 (nonmembrane spanning protein tyrosine kinase activity), and GO:0140096 (catalytic activity, acting on a protein). In CC, GO:0016021 (integral component of membrane), GO:0031224 (intrinsic component of membrane), and GO:0016020 (membrane) were significantly enriched. We further analyzed transcript changes using the Kyoto Gene and Genome (KEGG) pathway. Based on the KEGG pathway analysis, the REVs treatment in C. elegans mainly changed the metabolic pathway, peroxisome, and biosynthesis of cofactors pathway (Fig. 3F). In particular, metabolic pathway-related genes (ZK892.4, gst-8, gcy-20) and peroxisome-related genes (ZK892.4, F41E7.6) were upregulated by REVs, and biosynthesis of cofactor pathway-related genes (F17C8.9, sams-5, F52B11.2) was downregulated.
The top 5 genes increased by REV treatment are shown in Table 1. The expression of pas-3 (proteasome) and gnrr-1 (neuroactive ligand‒receptor interaction) were substantially increased. Among the five upregulated genes, substantially increased in genes related to proteasome (pas-3). Although not included in the top 5, genes associated with the immune response (F54D5.4, fbxa-116, sysm-1, irg-5, F55G11.2, tsp-1, C17H12.6, C34H4.1, Y43C5B.2, pals-23, T24C4.4, valv-1, C08E8.4) were significantly increased by REVs treatment in C. elegans. Conversely, REVs treatment significantly decreased the expression level of spe-1 (reproduction), arv-1 (lipid metabolic process), and sqd-1 (ortholog of human hepatocellular carcinoma) compared to SREVs treatment in C. elegans (Table 2).
DISCUSSION
Many studies are being conducted on the rumen of ruminants. The rumen microbiome is an essential component of the ruminant gastrointestinal tract, and it consists of bacteria (1010–1011 cells/mL), methanogenic archaea (107–109 cells/mL), protozoa (104–106 cells/mL), anaerobic fungi (103–106 cells/mL) and bacteriophages (109–1010 particles/mL) [2,30]. The rumen microbiome mainly acts as a fermenter and can digest ingested fibrin, which is not available to monogastric animals. Recently, research has been conducted to improve livestock production by understanding host–rumen microbial interactions. Studies focus on investigating the effect on rumen metabolism, methanogenesis, and host microRNA expression [30–33]. As the understanding between the host and the rumen has become more important, we attempted to identify the interaction with the host by isolating EVs released by various microorganisms present in the rumen.
Ruminal EVs were isolated using an ultracentrifuge, and particle shape and size were observed through TEM and NTA analysis. REVs were observed with various sizes ranging from 100–400 nm. This is a result of the release of EVs such as exosomes, microvesicles, microparticles and ectosomes through metabolic processes by various microorganisms in the rumen.
We used the C. elegans animal model to investigate the genetic changes of REVs on the host. REVs did not change the lifespan of C. elegans but increased the resistance of C. elegans to pathogenic bacteria. In transcriptome analysis, a significant increase in genes (gst-8, mtl-1) related to lifespan extension of C. elegans was not confirmed, so these findings are consistent with our findings. The C. elegans killing assay results showed that immune response genes (fbxa-116, sysm-1, irg-5, tsp-1, pals-23, and valv-1) were significantly increased. In particular, in previous studies, irg-5 is known as a necessary immune effector for host defense in pathogen infection and is a target of the p38 MAPK PMK-1 pathway [34,35]. Our findings demonstrate that treatment with REVs can stimulate C. elegans immune responses and induce more resistance to pathogenic bacteria.
According to the literature, early weaning of young ruminants has been reported to be essential for the rapid growth of ruminants, but early weaning can induce various stresses in young ruminants, which can cause growth retardation and suppression of the immune system [36,37]. Therefore, it is known that early settlement of microorganisms can be induced by transplanting rumen fluid to young ruminants for successful early weaning and rumen development, and digestive disorders of ruminants can be treated by transplanting rumen fluid [38,39]. Interestingly, consistent with our results, REVs upregulated the C. elegans membrane, developmental process, and structure-related genes in GO analysis. Similarly, according to KEGG analysis, treatment with REVs significantly upregulated the standard proteasome subunit pas-3 gene, which affects the embryonic viability, growth, fertility, and motility of C. elegans [35,40]. Particularly, upregulated chitinase activity by REVs treatment could indicate a relationship with increased host’s immune and defense functions. Chitinase activity can also reflect the increment of host digestibility, immunity, and protective systems, suggesting that it may play a role similar to previous studies of increasing the growth and development of young ruminants via rumen transplantation [41,42].
Currently, numerous continuing studies are elucidating the functionality and role of livestock-derived EVs in livestock industry fields [43–46]. As the first to isolate rumen derived EVs, this study showed that REVs increased pathogen resistance by regulating nematode immune genes. Furthermore, it could suggest that REVs may promote worm growth by significantly increasing genes involved in worm metabolism and development. Despite significant differences between ruminants and C. elegans, REVs stimulated genes involved in nematode metabolism and development, similar to the effects of ruminant transplantation in ruminants previously studied. These results suggest that REVs may bring growth and health benefits to ruminants. These data establish the basis for further investigation into the role of REVs. However, since these are the results of using the C. elegans model, additional studies are needed to deepen our understanding of the role of rumen EVs.
