Background
A tetraploid (4N) embryo complementation strategy has proven to be a useful model for post-implantation development of polyploid cells [1]. The 4N cells contribute to extra-embryonic tissues, but rarely contribute to the embryo itself [2,3]. In mice and cattle, the birth of chimeric offspring completely derived from embryonic stem (ES) cells or induced pluripotent stem (IPS) cells injected into or aggregated with 4N embryos has been reported [4–7]. However, there are apparently no reports of porcine chimeras constructed from ES- or IPS-like cells aggregated with 4N embryos. 4N embryos are usually produced by electrofusion of two-cell–stage embryos [8–10], inhibition of cleavage at an early stage [11], or by inhibition of extrusion of both first and second polar bodies [1,12,13]. It has proven to be very difficult to obtain 4N embryos through in vitro fertilization (IVF) owing to a high frequency of polyspermic fertilization events [12]. However, in the pig, the ploidy of 4N embryos produced using parthenogenetic embryos can more easily be predicted than that of 4N embryos obtained using IVF-derived embryos. This study was undertaken to compare the development, quality and ploidy transition of porcine 4N parthenogenetic embryos obtained through various methods.
Materials and Methods
All chemicals and reagents used in current study were purchased from Sigma Corporation (St. Louis, MO, USA), except where indicated. Animal experiments were approved by the Institutional Animal Care and Use Committee of Chungnam National University, Korea.
Freshly harvested porcine cumulus oocyte complexes (COCs) were rinsed 3–5 times and then cultured in four-well multidishes (50 oocytes/well) in 500 μL tissue culture medium (TCM) supplemented with 10% porcine follicular fluid (PFF), 10 ng/mL epidermal growth factor (EGF), 10 IU/mL pregnant mare serum gonadotropin (PMSG), and 10 IU/mL human chorionic gonadotropin (hCG) for 22 h. COCs were then cultured for an additional 22 h in the same IVM medium without hormones.
After a 44-h IVM period, oocytes were freed from follicle cells by treatment with 0.1% hyaluronidase. Denuded oocytes were washed several times in an activation solution (0.28 M D-mannitol, 0.1 mM MgSO4, 0.05 mM CaCl2, and 0.01% bovine serum albumin [BSA]), and then activated with a direct current pulse of 1,100 V/cm for 30 μs using an Electro Cell Manipulator 2001 (BTX, San Diego, CA, USA). Extrusion of the second polar body was inhibited by immediately culturing activated oocytes in Porcine Zygote Medium-3 (PZM-3) containing 3 mg/mL BSA plus 0 or 7.5 μg/mL cytochalasin B (CB) for 3 h, according to experimental requirements. After incubating with CB for 3 h, presumptive parthenogenetic embryos were transferred into PZM-3 and incubated at 38.5°C for 7 d in a 5% CO2 atmosphere. The day of activation was designated day 1. Cleavage rate (number of cleaved embryos/number of total tested embryos) and blastocyst formation rate (number of blastocysts/number of total tested embryos) were evaluated on culture days 3 and 7, respectively.
Click-iT EdU Imaging Kits (C10337; Invitrogen, Eugene, OR, USA) were used for EdU labeling according to the manufacturer’s instructions. Briefly, porcine 4N parthenogenetic embryos were cultured in EdU medium (10 μM) for 6 h at 38.5°C in a CO2 incubator. Embryos were fixed in 4% paraformaldehyde for 15 min and then permeabilized with 0.5% Triton X-100 for 15 min. Embryos were washed in 3% BSA, and then reacted with Click-iT reaction cocktail containing Alexa Fluor 488 azide. After EdU staining, samples were stained with 4’,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc., Burlingame, CA, USA) and examined under a confocal microscope.
Blastocyst stage embryos were collected and washed three times in PBS containing 0.1% polyvinyl alcohol (PBS-PVA). Blastocysts were then fixed and permeabilized with 4% paraformaldehyde and 0.5% Triton X-100 (v/v) for 1 h at room temperature. After washing several times with PBS-PVA, blastocysts were incubated with TUNEL reaction medium (In Situ Cell Death Detection Kit; TMR red; Roche, Germany) for 1 h at 38.5°C in the dark, and then counterstained with DAPI for 5 min in the dark to label all blastocyst nuclei. Finally, blastocysts were mounted and imaged under a fluorescence microscope.
4N parthenogenetic embryos derived from each experiment group were sampled and stored at −80°C until analysis. Expression levels of the pro-apoptotic gene BAX and anti-apoptotic gene BCL2L1 were examined using RT-PCR. Total RNA was isolated from each sample (20 blastocysts) using RNeasy Mini Kits (Qiagen, Valencia, CA, USA; Cat. No. 79254). cDNA was prepared using a TOPscript RT DryMIX kit (Enzynomics, Daejeon, Korea). RT-PCR was performed using an AccuPower PCR PreMix Kit (Bioneer, Daejeon, Korea) according to the manufacturer’s instructions. The primer pairs and annealing temperatures for the analyzed genes are listed in Table 1. The PCR products were separated by electrophoresis on 2% agarose gels containing EcoDye DNA Staining Solution (Biofact, Korea).
Chromosome analyses of embryos were performed using general procedures described previously, with modifications [13]. Embryos were collected and cultured for 6 h in PZM-3 medium containing 0.2 μg/mL colcemid, and then transferred to sodium citrate (0.8%, [w/v]) solution for 10–15 min. Individual embryos were then transferred onto glass slides and fixed with a methanol:acetic acid solution (1:1, [v/v]). After each glass slide had dried completely, chromosomes samples were stained with DAPI and coverslip-mounted. Chromosome numbers were determined under a fluorescence microscope.
After a 22-h IVM period, oocytes were treated with different concentrations of CB (0, 1, 5, 7.5, 15 μg/mL) for an additional 22 h. Oocytes were denuded completely and examined for the presence of PB1.
First, PB1 or non-PB1 oocytes were produced according to the needs of the experiment. For production of non-PB1 oocytes (Fig. 1A1-3 or Fig. 1C1-3), oocytes were matured without CB for 22 h, and then cultured with CB for an additional 22 h to inhibit PB1 extrusion. For production of PB1 oocytes (Fig. 1B1-3), oocytes were cultured without CB for 44 h to allow extrusion of PB1. Second, PB1 oocytes and non-PB1 oocytes were treated with or without CB for 3 h after electrical stimulation, and PB2 extrusion was monitored.
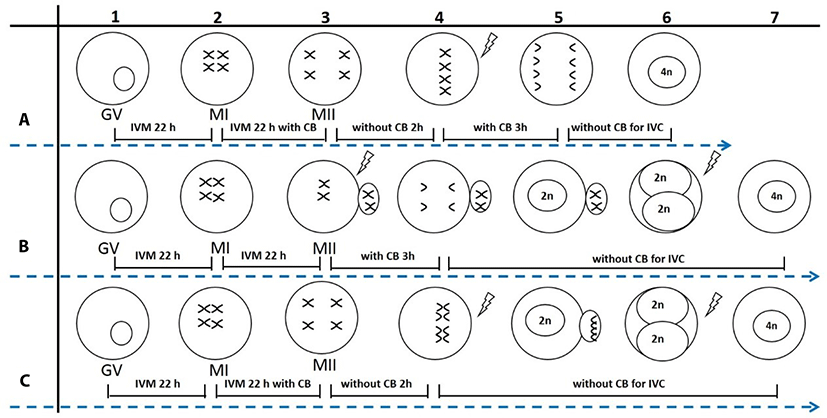
Porcine 2N parthenogenetic embryos were obtained by inhibition of PB2 (2N-I-PB2) or PB1 (2N-I-PB1) extrusion. For the 2N-I-PB2 method (Fig. 1B3-5), PB1 oocytes were activated and then treated with CB for 3 h to inhibit PB2 extrusion. For the 2N-I-PB1 method (Fig. 1C3-5), non-PB1 oocytes were activated and cultured without CB to allow PB2 extrusion. Finally, 2N parthenogenetic embryos were cultured in PZM-3 medium, and cleavage and blastocyst formation rates were investigated on culture days 3 and 7, respectively.
2-Cell stage 2N parthenogenetic embryos derived from 2N-I-PB2 (Fig. 1B6) or 2N-I-PB1 (Fig. 1C6) groups were pre-equilibrated in fusion solution (same as electro-activation solution), and then embryos in the chamber were aligned with their fusion plane parallel to the two electrodes. Fusion was achieved with a direct current pulse of 1,100 V/cm for 30 μs using an Electro Cell Manipulator. Embryos were observed at 30 min post electrofusion, and fused embryos were cultured in PZM-3 for further study. 2-Cell embryos derived from 2N-I-PB2 and 2N-I-PB1 groups that were not subjected to electrofusion were used as control groups (C-2N-I-PB2 and C-2N-I-PB1).
Porcine 4N parthenogenetic embryos were produced by inhibition of both PB1 and PB2 extrusion, termed Method 1 (M1), or by electrofusion of 2-cell stage diploid parthenogenetic embryos derived from 2N-I-PB2 (M2) or 2N-I-PB1 (M3). For M1, non-PB1 oocytes (Fig. 1A3) were activated and then treated with CB for 3 h to inhibit PB2 extrusion (Fig. 1A3-6). After treating with CB for 3 h, 4N embryos were produced by selecting and culturing non-PB1/non-PB2 embryos in CB-free medium. For M2, completely fused 2-cell stage 2N embryos derived from 2N-I-PB2 (Fig. 1B7) were used to produce 4N embryos. For M3, completely fused 2-cell stage 2N embryos derived from 2N-I-PB1 (Fig. 1C7) were obtained to produce 4N embryos. Finally, 4N parthenogenetic embryos from each group were cultured in PZM-3 medium. Cleavage and blastocyst formation rates were investigated on culture days 3 and 7, respectively. Total and EdU-positive cell numbers in blastocysts were also evaluated using EdU labeling.
Data were analyzed by one-way analysis of variance (ANOVA) followed by Fisher’s protected least significant difference test or t-test using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Percentage data were subjected to arcsine transformation before analysis. At least three replicates were performed for each experiment, and results are expressed as means ± standard error (se). p-values < 0.05 were considered significant.
Results
As shown in Fig. 2A, there was no difference in the rate of PB1 extrusion between control and 1 μg/mL CB groups. However, the rate of PB1 extrusion significantly decreased with further increases in CB concentration compared with control and 1 μg/mL groups, plateauing at approximately 7.5 μg/mL. Based on these results, we used 7.5 μg/mL CB for subsequent studies.
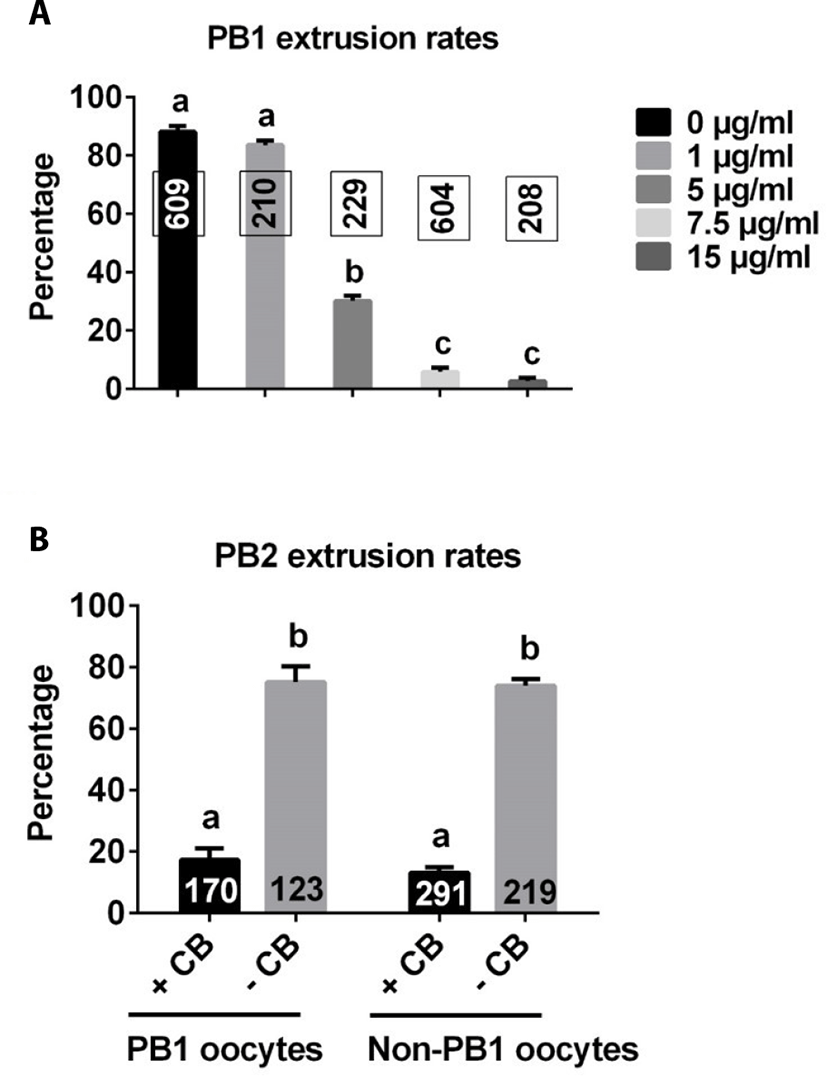
PB1 embryos and non-PB1 embryos were treated with CB (7.5 μg/mL) for 3 h after electrical stimulation. The rate of PB2 extrusion was significantly lower with CB treatment than without CB treatment in both PB1 oocytes and non-PB1 oocyte groups (Fig. 2B).
There were no differences in the rates of cleavage or blastocyst formation, or total cell numbers in 2N parthenogenetic blastocysts between 2N-I-PB2 and 2N-I-PB1 groups (Fig. 3A, B).
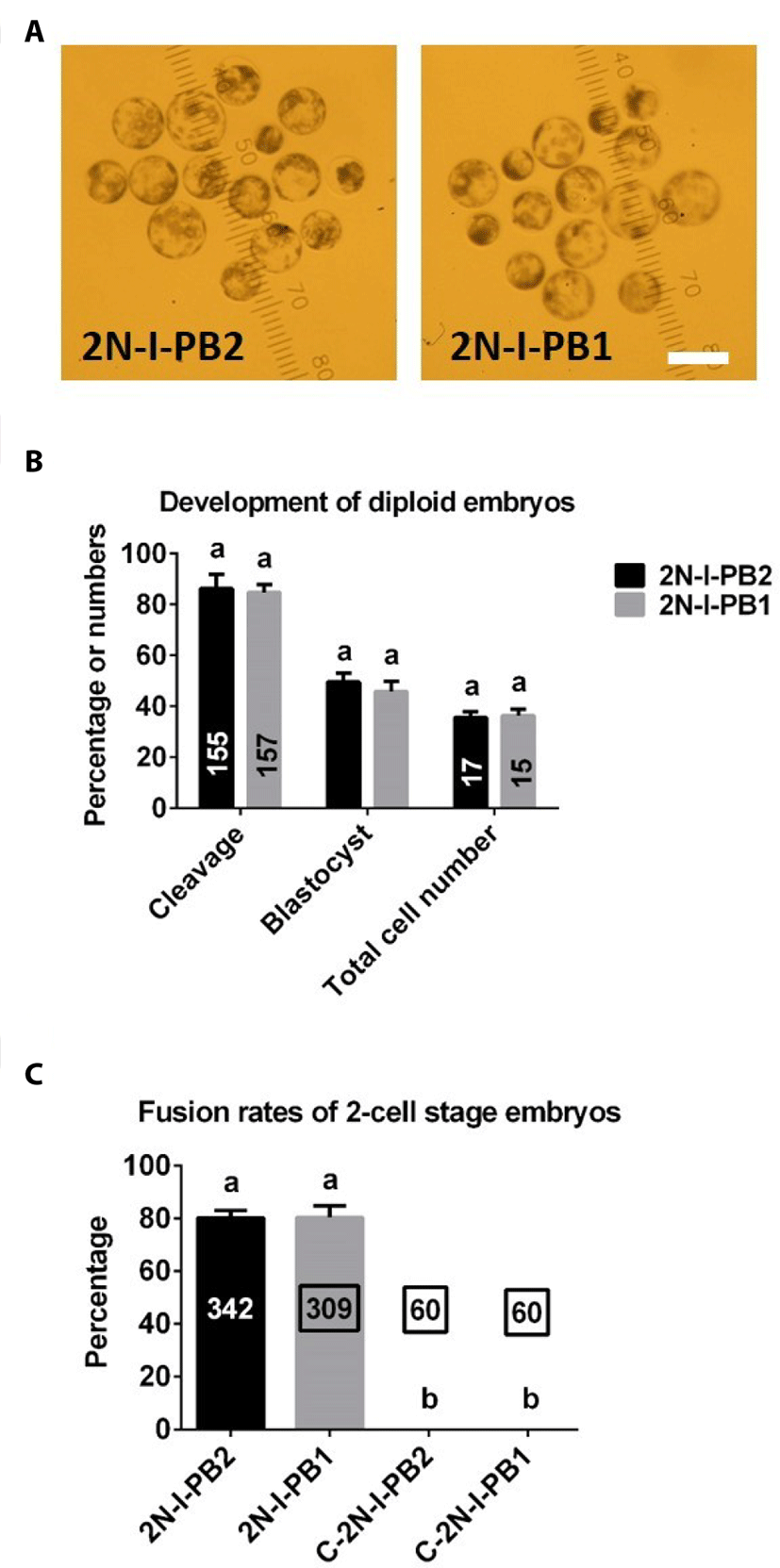
As shown in Fig. 3C, there were no significant differences in electrofusion rates between 2N-I-PB2 and 2N-I-PB1 groups. As expected, no fusion occurred in C-2N-I-PB2 and C-2N-I-PB1 control groups (Fig. 3C).
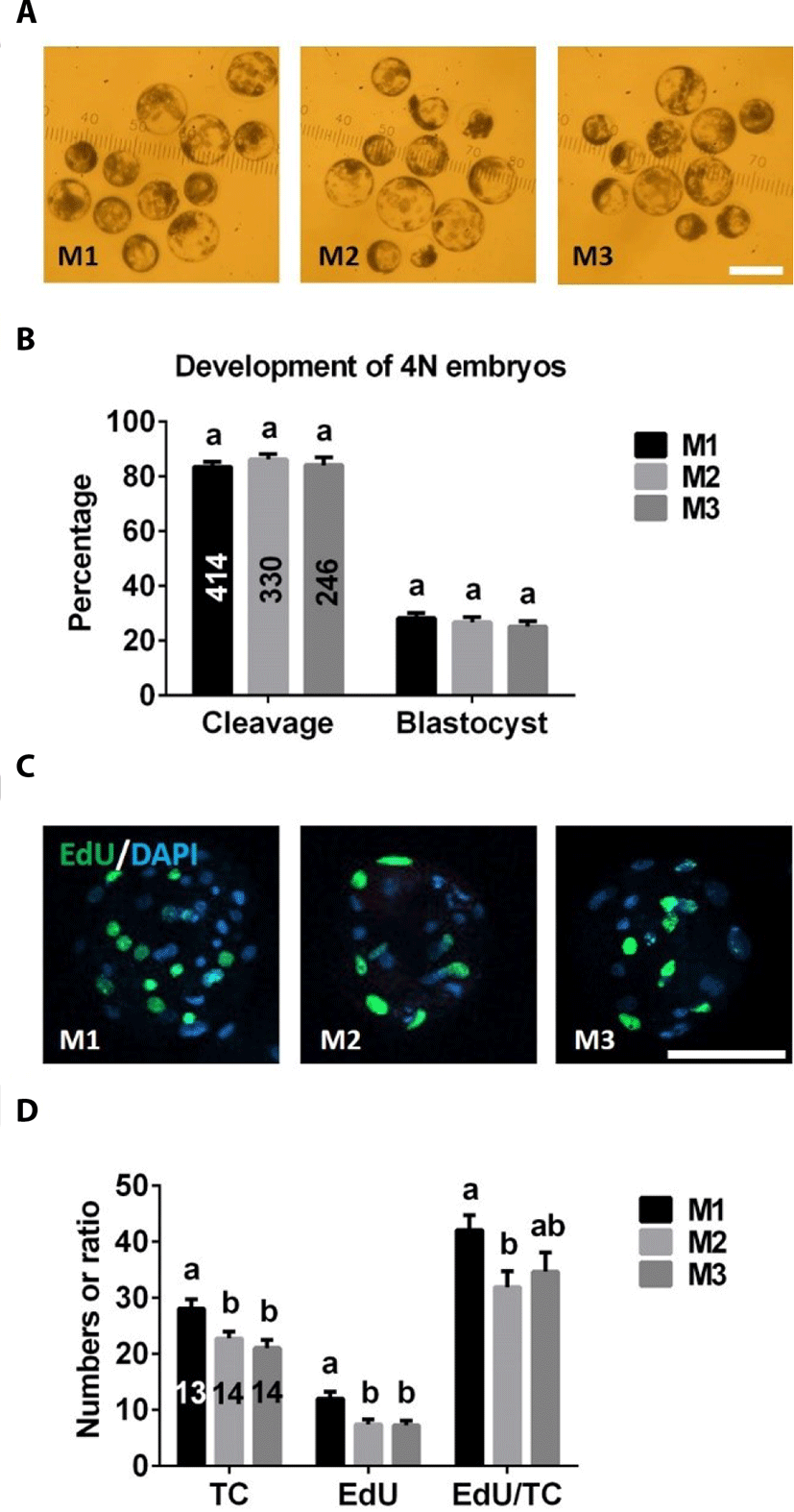
There were no significant differences in cleavage or blastocyst formation rates among M1, M2, and M3 groups (Fig. 4A, B). However, total and EdU-positive cell numbers of 4N blastocysts were significantly higher in the M1 group than in M2 or M3 groups (Fig. 4C, D). There was no difference in the ratio of EdU-positive cell numbers to total cell numbers between M2 and M3 groups, but the value of this ratio was significantly lower in the M2 group than the M1 group (Fig. 4D).
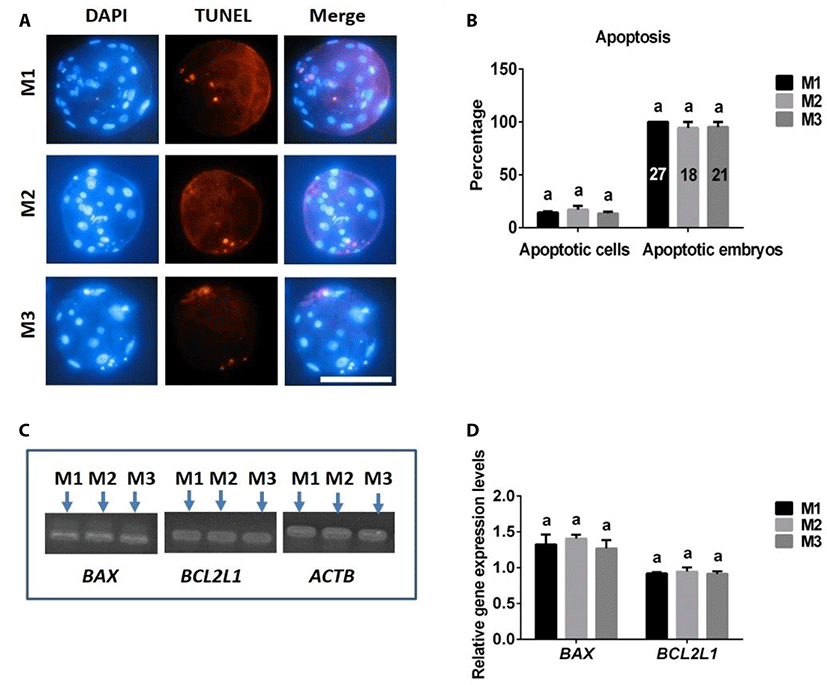
The frequencies of apoptotic cells in blastocysts and embryos were similar in 4N blastocysts produced with M1, M2, and M3 (Fig. 5A, B). There were no differences in expression levels of the pro-apoptotic BAX gene or anti-apoptotic BCL2L1 gene among embryos derived from M1, M2, and M3 groups (Fig. 5C, D).
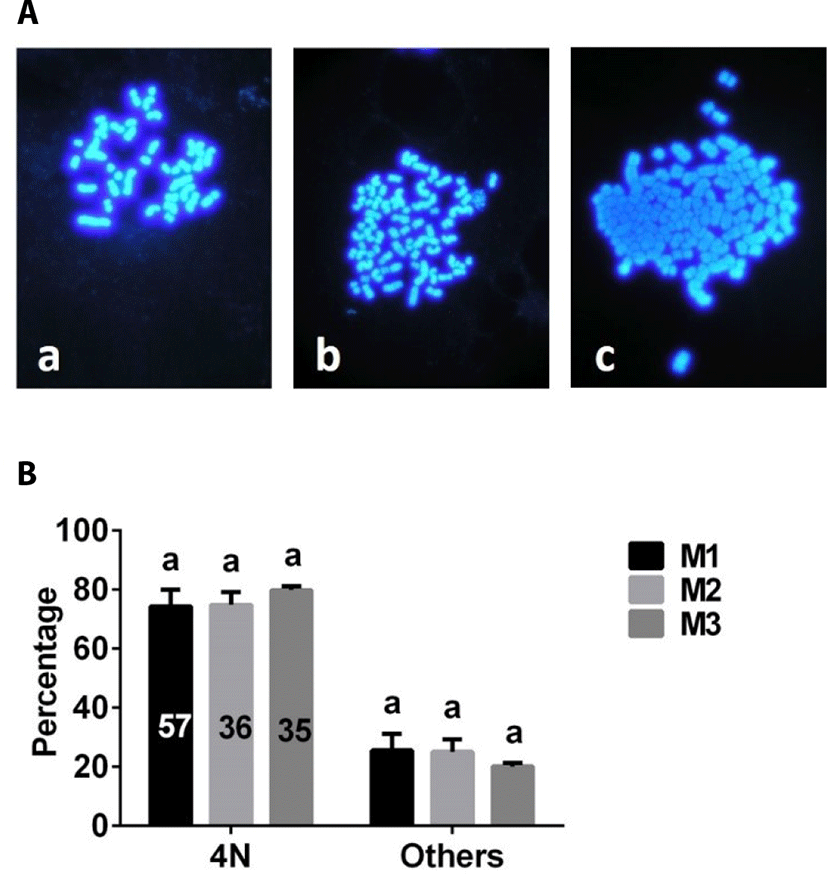
The number of chromosome was analyzed after chromosome staining (Fig. 6A). There were no differences in the proportion of 4N embryos among M1, M2, and M3 groups (Fig. 6B).
Discussion
Here, we compared the effectiveness of a variety of methods for producing porcine 4N parthenogenetic embryos. Our results indicated that porcine 4N parthenogenetic embryos could be produced using each of the methods tested. However, porcine 4N embryos derived from M1 contained more nuclei than those of embryos derived from M2 or M3, suggesting that M1 is superior to M2 and M3. Because the physiological characteristics of parthenogenetic embryos are similar to those of fertilized embryos, production of parthenogenetic embryos is important in studies of early preimplantation embryo development [14]. Development of parthenogenetic embryos has been extensively evaluated in many mammalian species. In general, the developmental competence of 2N embryos is more robust than that of haploid (1N) or 4N embryos [15,16]. In the current study, the blastocyst formation rates of 2N embryos (Fig. 3B) derived from inhibition of PB2 (2N-I-PB2 group) and PB1 (2N-I-PB1 group) extrusion methods were clearly higher than those of 4N embryos (Fig. 4B) derived from M1, M2, or M3. Our results are consistent with previous reports that the developmental potential of pig [16] and bovine [1] 4N parthenogenetic embryos is compromised compared with that of 2N parthenogenetic embryos. The similarity of both the development and ploidy of porcine 4N parthenogenetic embryos derived from M1, M2, and M3 suggests that these methods are equivalent for producing porcine 4N parthenogenetic embryos. In the current study, a chromosomal number counting analysis was used to evaluate the ploidy of porcine parthenogenetic embryos. If ploidy of embryos were evaluated using fluorescence in situ hybridization (FISH) analyss [17], the results might be more precise.
The total cell number in blastocysts is a very important parameter for evaluating the quality of embryos [18]. Usually, 2N blastocysts have more nuclei than 4N embryos [12,16,19]. In the present study, we found that, although the morphology of 4N and 2N parthenogenetic blastocysts was similar, the total cell number in 4N blastocysts (Fig. 4D) was lower than that in 2N embryos (Fig. 3B). However, total cell numbers in 4N parthenogenetic blastocysts derived from M1 were significantly higher than those from M2 and M3. This result was not surprising since it is known that the 4N embryos were produced by blastomere fusion, halting one cell division; thus, 4N embryo-derived blastomere fusion methods (M2 and M3) lost one cleavage compared with that of 4N embryos derived from inhibition of PB1 and PB2 (M1). In addition, we also found that EdU-positive cell numbers were significantly higher in 4N blastocysts derived from the M1 group than the other two groups, indicating that 4N parthenogenetic embryos obtained by inhibiting PB1 and PB2 (M1) possess more powerful cell proliferation potential.
In addition to cell numbers, apoptosis is another important indicator of embryo quality [20,21]. Using TUNEL assays and RT-PCR analyses to assess apoptosis in porcine 4N parthenogenetic embryos, we found no differences in apoptosis rate (TUNEL-positive cells/total cells) or expression levels of the anti-apoptotic BCL2L1 gene or pro-apoptotic BAX gene among blastocysts produced from M1, M2, and M3, suggesting that apoptosis was not influenced by the different 4N parthenogenetic embryo-generation methods.
In conclusion, we report the production of porcine 4N parthenogenetic embryos by inhibition of both PB1 and PB2 (M1), or electrofusion of 2-cell stage 2N parthenogenetic embryos derived from inhibition of PB2 (M2) or PB1 (M3) extrusion. Total and EdU-positive cell numbers in 4N parthenogenetic embryos derived from the M1 group were significantly higher than those from M2 and M3 groups, although in each case embryos had similar developmental potential and 4N ploidy proportion. Different methods also did not change apoptosis rates in 4N parthenogenetic embryos. Our study provides an efficient approach for producing 4N parthenogenetic embryos, and establishes reference values for the production and development of porcine 4N parthenogenetic embryos.
