RESEARCH ARTICLE
Complete genome sequence of Lactococcus lactis strain K_LL005, a xylose-utilizing bacterium isolated from grasshopper (Oxya chinensis sinuosa)
Hyeri Kim
1,#
, Robin B. Guevarra
1,#
, Jae Hyoung Cho
1,#
, Hyeun Bum Kim
1,*
, Ju-Hoon Lee
2,*
1Department of Animal Resources Science, Dankook University, Cheonan 31116, Korea
2Department of Food and Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul 08826, Korea
*Corresponding author: Hyeun Bum Kim, Department of Animal Resources Science, Dankook University, Cheonan 31116, Korea. Tel: +82-41-550-3653, E-mail:
hbkim@dankook.ac.kr
*Corresponding author: Ju-Hoon Lee, Department of Food and Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul 08826, Korea, Tel: +82-2-880-4854, E-mail:
juhlee@snu.ac.kr
#These authors contributed equally to this work.
© Copyright 2021 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Sep 10, 2020; Revised: Nov 06, 2020; Accepted: Nov 10, 2020
Published Online: Jan 31, 2021
Abstract
Lactococcus lactis is a fermentative lactic acid bacterium that is used extensively in food fermentations. The L. lactis strain K_LL005 was isolated from the grasshopper (Oxya chinensis sinuosa) gut in Korea. In this study, we reported the complete genome sequence of Lactococcus lactis K_LL005. The final complete genome assembly consist of one circular chromosome (2,375,093 bp) with an overall guanine + cytosine (G + C) content of 35.0%. Annotation results revealed 2,281 protein-coding sequences (CDSs), 19 rRNAs, and 68 tRNA genes. Lactococcus lactis K_LL005 has a gene encoding xylose metabolism such as xylR, xylA, and xylB (xylRAB).
Keywords: Lactococcus lactis strain K_LL005; Grasshopper; Whole genome sequencing; Xylose
In nature, Lactococcus lactis was originally isolated from plant or animal surfaces, where it was known to be dormant, and only actively multiplying in the intestinal tract after being consumed by a ruminant [1]. For the past 20 years, L. lactis has extremely extended its application from food to being a successful microbial industry. As a result, the species of L. lactis has been domesticated for centuries in the fermentation of food, especially cheese and yoghurt [2].
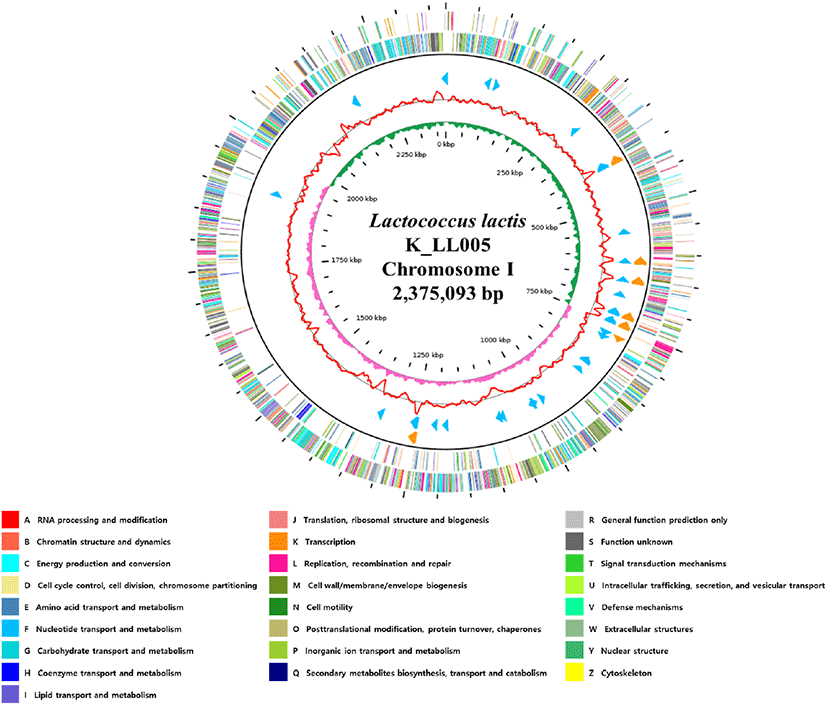
In the present study, the L. lactis strain K_LL005 was isolated from the gut of a grasshopper (Oxya chinensis sinuosa) collected from local farm in Yangyang, Gangwon-do, Korea. The L. lactis K_LL005 was grown in de Man-Rogosa-Sharpe broth at 37°C for 24 h. Genomic DNA was extracted using the MagAttract HMW DNA Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. The complete genome of the L. lactis K_LL005 was sequenced using the PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) platform at Insilicogen (Yongin, Korea). Library preparation was performed using SMRTbell™ Template Prep Kit 1.0 following the manufacturer’s instructions (Pacific Biosciences). PacBio sequencing resulted in 798,317,798 base pairs and 109,112 long reads after filtering. The sequence reads were de novo assembled using the hierarchical genome assembly process (HGAP v2.3.0) workflow, and further polished with Quiver. Because bacterial genomes and plasmids are typically circular, we checked the forms for each of contigs using MUMmer 3.5 [3] and trimmed one of the self-similar ends for manual genome closure. Putative protein coding sequences were predicted using Glimmer v3.02 [4], rapid prokaryotic genome annotation (PROKKA) v1.14.5 [5] and Rapid Annotation using Subsystem Technology (RAST) v2.0 [6]. The results from the three prediction methods were confirmed by manual curation [7]. The genes of rRNA and tRNA were predicted using RNAmmer v1.2 and tRNAscan-SE v1.4, respectively [8,9]. GO annotation was assigned to each of ORFs using Blast2GO software [10] to analyze the best hits of the BLAST results. The complete genome of the L. lactis strain K_LL005 contains one circular chromosome (2,375,093 bp) with a guanine + cytosine (GC) content of 35.0%, 2281 predicted open reading frames (ORFs), 19 rRNA genes, and 68 tRNA genes. The genome feature and map of L. lactis strain K_LL005 are illustrated in Table 1 and Fig 1.
Table 1.
Genome features of Lactococcus lactis strain K_LL005
|
Property |
Term |
|
Libraries used |
PacBio SMRTbell™ library |
|
Sequencing platforms |
PacBio RS II sequencer |
|
Assemblers |
PacBio SMRT analysis v2.3.0 HGAP.3 |
|
Annotation method |
Glimmer v3.02, PROKKA v1.14.5 and RAST v2.0 |
|
Average genome coverage |
267x |
|
Chromosome length (bp) |
2,375,093 bp |
|
No. of contigs |
1 |
|
Guanine + cytosine (G + C) content (%) |
35.0 |
|
Protein–coding genes (CDSs) |
2,281 |
|
rRNA genes |
19 |
|
tRNA genes |
68 |
|
Plasmids |
0 |
|
Genbank Accession No. |
CP060580 |
Download Excel Table
Fig. 1.
Genome map of Lactococcus lactis strain K_LL005.
The outer circle denotes the locations of all annotated ORFs, and the inner circle with the red denotes GC content. Pink and green peaks denote GC skew. The orange arrows denote rRNAs, and the sky blue arrows denote the tRNA operons. All annotated ORFs are colored differently based on the COG assignments. G, guanine; C, cytosine; ORFs, open reading frames; COG, clusters of orthologous groups.
Download Original Figure
The genome of L. lactis K_LL005 possessed xylA, xylB, and xylR (xylRAB) encoding enzymes and protein for xylose utilization. Xylose is a component of hemicellulose, an important structural polysaccharide in plants, and xylose utilization metabolism required the products of the xylRAB genes encoding putative positive transcriptional regulator, xylose isomerase, and xylulose kinase. Furthermore, L. lactis K_LL005 has the genes, MnnB, RbsK, and FrcK, which are associated with mannose utilization, D-ribose utilization and fructose utilization.
Acknowledgements
We thank Mo Re Kim (Brandeis University, Waltham, MA, USA) for the English grammar corrections.
Availability of data and material
Ethics approval and consent to participate