INTRODUCTION
Porcine circovirus type 2 (PCV2), a small non-enveloped DNA virus, is perceived as an important pathogen in the swine industry worldwide [1]. PCV2 has four major open reading frames (ORFs). ORF1 encodes the replicase (Rep) and splicing replicase (Rep’) proteins that participate in viral replication [2,3]. ORF2 encodes a viral capsid protein (Cap) [4,5]. The ORF3-encoded protein has been reported to be involved in the apoptosis of PCV2 infected cells [6]. The fourth ORF, named ORF4, has also been reported as a mitochondrial-targeting protein that interacts with adenine nucleotide translocase 3 via the mitochondrial pathways [7].
The proliferation of virus titers correlates with weight loss in post-weaning piglets [8,9]. The typical symptoms of PCV2 associated disease (PCVAD) are an increase in viral load and a decrease in average daily gain (ADG) [10-12]. Moreover, the appearance of PCVAD in swine herds leads to low productivity and economic losses for those farms [13-15]. In addition, PCVAD is known to be influenced by the host genetic background as to which genetically resistant or susceptible to PCV2 [16]. Therefore, the control of PCV2 viral load through host genetic effects is considered important.
Regulator of G protein signaling 16 (RGS16), a GTPase-activating protein, can regulate T-lymphocyte activation [17]. It has previously been reported that RGS16 is upregulated in inflammatory dendritic cells stimulated with lipopolysaccharide [18]. Furthermore, RGS16 is associated with the ORF3 protein of PCV2 and host response [19]. In pigs, the RGS16 gene is located in the quantitative trait loci for ADG on Sus scrofa chromosome 9 [20,21]. Accordingly, RGS16 is thought to be associated with PCVAD. The aim of this study was to identify polymorphisms in the RGS16 region and investigate the association between genotypes and the PCV2 viral load. Four-week-old unvaccinated piglets against PCV2 were used for measuring PCV2 viral load by blood collection. After 6 weeks, the viral load of PCV2 was measured again via re-blood collection.
MATERIALS AND METHODS
This study was conducted in accordance with the animal care guidelines of the Institutional Animal Care and Use Committee of the National Institute of Animal Science, Korea.
A total of 142 unvaccinated Yorkshire pigs (65 castrated males and 77 females) which were born from the 46 different sows selected from a single farm were chosen to have zero viral load at 4 weeks of age and were naturally infected with PCV2. Peripheral blood was collected from each pig at 4 and 10 weeks of age to assess PCV2 viral load and genomic DNA.
PCV2 viral DNA was extracted from serum using the QIAGENTM virus mini elute kit (QIAGEN, MD, USA). PCV2 viral load was measured using TaqManTM probe quantitative polymerase chain reaction (qPCR) with the primers listed in Table 1. These primers were designed from the complete PCV2 genome (Korea, GenBank: FR823451.1) using Primer3 [22]. The reaction mixture contained 500 nM of each primer, 250 nM probe, 25 μL of 2× TaqManTM Universal Master Mix (Applied Biosystems, Waltham, MA, USA), and 2.5 μL template. Nuclease-free water was added to bring the final volume to 50 μL. Amplification was performed under universal cycling conditions (2 min at 50°C, 10 min at 95°C, and 45 cycles of 15 s at 95°C and 1 min at 58°C).
To identify polymorphisms, between or within four different pig breeds (Yorkshire, Landrace, Duroc, and Berkshire; n = 20 per breed), direct sequencing analysis was performed using porcine-specific primers containing the upstream and genetic regions of porcine RGS16. The primer sets were designed using the Primer3 software [22] (Table 2). PCR was performed using a MastercyclerTM gradient (Eppendorf, Hamburg, Germany) in a total volume of 20 μL containing 100 ng porcine genomic DNA template, 10 pmol of each primer, 0.25 mM of each dNTP, 1× PCR buffer, and 1.25 U DNA polymerase (i-Max II, Intron biotechnology, Seongnam, Korea). Sequencing analysis was performed using an ABI PRISM 3730 Genetic Analyzer (Applied BiosystemsUSA). Sequence assembly and polymorphism identification were executed using SeqMan [23].
Genotyping was performed on the rs332913874, rs326071195, and rs318298586 loci. Genomic DNA was extracted from peripheral blood using a DNA isolation kit (G-DexTM IIb, Intron Biotechnology). These single nucleotide polymorphism (SNP) genotypes were identified via direct sequencing analysis. PCR was performed using the aforementioned conditions. The amplification conditions were as follows: 94°C for 10 min; 30 cycles of 94°C for 1 min, 61°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 10 min.
The significance of differences in genotype and allele frequencies within the population was determined using the Hardy–Weinberg equilibrium (HWE) calculation (chi-square test). The association of SNPs with viral load was evaluated using the MIXED model procedure in the SAS software (ver. 9.4, SAS Institute, Cary, NC, USA). The model was used as follows: yijkl = μ + Si + Bj + Dk + eijkl, where yijklmis the observation of traits, μ is the general mean, Si is the fixed effect of SNP genotypes i, Bj is the fixed effect of batch j, Dk is the random effect of dam l, and eijkl is the random error. Haplotype combinations of three SNPs were reconstructed by PHASE software [24] and analyzed with the same MIXED procedure using SAS. Results are presented as the mean of the least squares with standard error. Additive and dominance effects were calculated using the method previously described by Hasenstein et al. [25]. A value of p < 0.05 was considered statistically significant.
RESULTS
We identified 22 SNPs in the upstream and genetic regions of RGS16 inYorkshire pigs (Fig. 1). Of these, 10 SNPs, including rs332913874 and rs326071195, were located in the upstream region, whereas 12 others, including rs318298586, spread over the intron region. These 22 SNPs were registered on dbSNP and are presented in Supplementary Table S1.
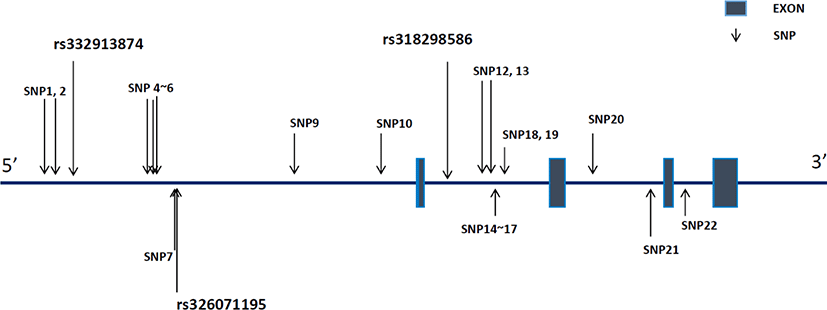
The allelic and genotypic frequencies of rs332913874, rs326071195, and rs318298586 areshown in Table 3. In rs332913874, the G and A allele frequencies were 0.64 and 0.36, respectively, in the Yorkshire population. Genotype frequencies for the homozygote (GG and AA) and heterozygote (GA) genotypes were 0.38, 0.10, and 0.52, respectively. The genotype and allele frequencies of rs326071195 were found to be as follows: T allele, 0.52; C allele, 0.48, TT genotype, 0.19; TC genotype, 0.67; and CC genotype, 0.14. Furthermore, the genotype and allele frequencies of another SNP (rs318298586) were as follows: A allele, 0.51; T allele, 0.49; AA genotype, 0.19; AT genotype, 0.65; and TT genotype, 0.16. Chi-square tests were used to check for deviation from the HWE. We found that the rs332913874 genotypes conformed to HWE; however, the genotypes of the other two SNPs did not follow HWE in our population. Based on these frequency data, we conducted an association analysis between genotypes and the PCV2 viral load in our population.
Haplotype analysis of the three SNPs is presented in Table 4. We observed three major haplotypes that accounted for 97.5% of all possible haplotype combinations in the population of animals. Based on these three haplotype combinations (Haplo1, Haplo4, and Haplo6), we performed association analysis with haplotype combinations and the PCV2 viral load. Results of the analyses are presented in Table 5. The Haplo1 (GTA) and Haplo6 (ACT) combinations showed significant differences in PCV2 viral loads. Animals containing one copy of the Haplo1 combination had a significantly higher PCV2 viral load than those containing two copies (p = 0.04); However, compared to that of a single copy of Haplo1, the absence of Haplo1 combination did not significantly affect PCV2 viral load. The Haplo4 (GCT) combination did not significance. The diplotype frequencies of haplotpype combinations were showed in Supplementary Table S2. The frequency of the Haplo1/Haplo6 (GTA/ACT) combination which had the most count was 0.465. The second and the third count of diplotype combinations were Haplo1/Haplo1 (GTA/GTA) and Haplo1/Haplo4 (GTA/GCT). The frequencies of these two combination were 0.176 and 0.169 each. The forth frequency of diplotype combinations (Haplo6/Haplo6, ACT/ACT) was 0.099. The associations among the top 4 diplotype combinations against viral load were presented in Supplementary Table S3. The viral load of Haplo1/Haplo6 (GTA/ACT) combination was significantly higher than Haplo1/Haplo1 (GTA/GTA) combination (p = 0.01).
The animal frequency against viral load at 10-week old is showed using a histogram in Supplementary Fig. S1. The interval of the viral load was set by positive integer values. The results of the analyzed associations between the genotypes of SNPs in RGS16 and PCV2 viral loads are presented in Table 6. The rs332913874 SNP was significantly associated with PCV2 viral load (p = 0.03). Animals with the heterozygous genotype had a higher viral load than those with the homozygous GG genotype. However, the heterozygous GA genotype did not show any statistical significance with another homozygous AA genotype. The association analysis of rs326071195 and PCV2 viral load revealed that the viral load in animals with the TC genotype was higher than that in animals with the TT genotype; however, no significance was observed between the viral loads in animals with the CC and heterozygous genotypes. The results of residual normality test are showed in Supplementary Fig. S2. All of the residual was satisfied normality. In the case of rs318298586, the viral load of pigs with the AT genotype was higher than that of pigs with the AA genotype. Conversely, no significant difference against viral load was found between the heterozygous and TT genotypes.
DISCUSSION
In this study, we investigated the association between PCV2 viral load and SNP genotypes in porcine RGS16. PCV2, a small non-enveloped, circular ssDNA virus, leads to PCVAD with clinical signs of wasting disease, enlarged lymph nodes, and diarrhea [9,11,16,26]. PCVAD is an endemic disease that causes reduced productivity in the swine industry [1,10,27]. PCV2 is commonly known to have 11 ORFs [16,28]; however, the functions of only four of these ORF proteins have been identified. ORF1 codes for nonstructural replicase proteins known as Rep and Rep’ [5,29]. ORF2 encodes the Cap protein [30,31]. ORF3 proteins have been previously reported to induce apoptosis in PK-15 cells [32]. ORF4 is known to be a mitochondrial-targeting protein that triggers apoptosis in the host cell [7].
Various types of research on PCV2 have revealed that the viral load of experimental or naturally infected pigs is associated with PCVAD clinical signs, including postweaning multisystemic wasting syndrome (PMWS) [33-36]. To prevent PCVAD, vaccinations against PCV2 are done in the swine farms. Nevertheless, the swine industry has been consistently damaged by PCV2. Therefore, researchers have experimented on host genetic susceptibility to PCV2 using infection and genome-wide association studies [37,38]. Walker et al. reported that porcine SYNGR2 polymorphism is related to the PCV2 viral load. Zhang et al. also reported that some miRNAs are associated with PCVAD susceptibility to PCV2 infection in lung tissues. In this study, we investigated the differences in viral load based on genotype in the RGS16 polymorphism in naturally infected Yorkshire pigs. We identified SNPs in porcine RGS16 gene. A total of 22 SNPs were identified, including those in the 5′UTR and 3′UTR regions. In the statistical analysis model, we did not include sex as a fixed effect. There was no significant effect between sex in the statistical model including sex as a fixed effect. The previous study reported that the difference of sex effect against PCV2 was not founded [39]. Therefore, it is estimated that there was no difference in infection with gender in our study. Three SNPs located in the intron region were associated with the PCV2 viral load. In particular, the heterozygous genotypes of these three SNPs were related to high PCV2 viral loads. Furthermore, the reference homozygous genotypes (Allele1/Allele1) were associated with a low viral load. In addition, the association between SNP and viral load did not follow the additive effects and the complete dominance effect. This phenomenon is presumed to be the effect of hybrid weakness, but more follow-up studies seem to be needed. As previously described, PCV2 viral load is a triggering factor that causes PCVAD, including PMWS. Moreover, RGS16 has been previously reported to be involved in the activation of immune cells, with the important exception of macrophages in rats [40,41]. Another study has reported that RGS16 activated by inflammatory stimulation and the PCV2 ORF3 protein colocalize in porcine peripheral blood mononuclear cells [19]. In the present study, we did not identify differences in the expression levels of RGS16 in porcine whole blood among the genotypes. In addition, the PCV2 viral load was not measured every week in the study animals; hence, it was difficult to trace the change of aspect against viral load from the initial point to the end point.
CONCLUSION
Although the measurement of viral load in this study showed two points, our findings confirm that the polymorphisms in the RGS16 gene have considerable effects on the PCV2 viral load in naturally infected pigs. Therefore, these polymorphisms can be regarded as an effective marker for estimating the proliferation of PCV2 viral load.
