RESEARCH ARTICLE
Complete genome sequence of Lactococcus taiwanensis strain K_LL004, encoding hydrolytic enzymes of plant polysaccharides isolated from grasshopper (Oxya chinensis sinuosa)
Hyunok Doo
1,#
, Hyeri Kim
1,#
, Jin Ho Cho
2,#
, Minho Song
3,#
, Eun Sol Kim
1
, Jae Hyoung Cho
1
, Sheena Kim
1
, Gi Beom Keum
1
, Jinok Kwak
1
, Sriniwas Pandey
1
, Hyeun Bum Kim
1,*
, Ju-Hoon Lee
4,*
1Department of Animal Resources Science, Dankook University, Cheonan 31116, Korea
2Department of Animal Science, Chungbuk National University, Cheongju 28644, Korea
3Division of Animal and Dairy Science, Chungnam National University, Daejeon 34134, Korea
4Department of Food Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul 08826, Korea
# These authors contributed equally to this work.
*Corresponding author: Hyeun Bum Kim, Department of Animal Resources Science, Dankook University, Cheonan 31116, Korea. Tel: +82-41-550-3653, E-mail:
hbkim@dankook.ac.kr
*Corresponding author: Ju-Hoon Lee, Department of Food Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul 08826, Korea. Tel: +82-2-880-4854, E-mail:
juhlee@snu.ac.kr
© Copyright 2023 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: Sep 07, 2022; Revised: Oct 13, 2022; Accepted: Oct 31, 2022
Published Online: May 31, 2023
Abstract
The Lactococcus taiwanensis strain K_LL004 was isolated from the gut of a grasshopper (Oxya chinensis sinuosa) collected from local farm in Korea. L. taiwanensis strain K_LL004 is the functional probiotic candidate with an ability to hydrolyse plant polysaccharides. The complete genome of the L. taiwanensis strain K_LL004 contains one circular chromosome (1,995,099 bp) with a guanine + cytosine (GC) content of 38.8%. Moreover, 1,929 Protein-coding sequence, 19 rRNA genes, and 62 tRNA genes were identified based on results of annotation. L. taiwanensis strain K_LL004 has a gene, which encodes hydrolytic enzymes such as beta-glucosidase and beta-xylosidase, that hydrolyzes plant polysaccharides.
Keywords: Lactococcus taiwanensis; Grasshopper; Beta-glucosidase; Beta-xylosidase; Whole genome sequencing
Lactococcus is a genus of lactic acid bacteria (LAB) that are present on grass and other plant material and in the gastrointestinal tracts [1]. Twenty-two species of the genus Lactococcus are established till date. In particular, Lactococcus lactis is the most common strain which is used as a starter in food fermentation [2]. Lactococcus taiwanensis, a type of Lactic acid Bacteria, has not been studied in detail, and therefore the genomic information of Lactococcus taiwanensis is limited.
In the present study, the L. taiwanensis strain K_LL004 was isolated from the gut of a grasshopper (Oxya chinensis sinuosa), an insect preferring to feed on plants, collected from local farm in Yangyang, Gangwon-do, Korea. The L. taiwanensis strain K_LL004 was grown in de Man-Rogosa-Sharpe broth at 37°C for 24 h. Genomic DNA was extracted using the MagAttract HMW DNA Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. The complete genome of the L. taiwanensis strain K_LL004 was sequenced using the PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) platform at Insilicogen (Yongin, Korea). Library preparation was performed using SMRTbell™ Template Prep Kit 1.0 following the manufacturer’s instructions (Pacific Biosciences). PacBio sequencing produced 161,058 of long reads and 1,143,521,995 base pairs after subreads filtering. De novo assemble was performed using the hierarchical genome assembly process (HGAP v2.3.0) workflow and polished using Quiver. The quality of genome assembly was assessed by using Quality Assessment Tool for Genome Assemblies (QUAST) v5.0.2 [3]. The quantitative assessment of the genome completeness was conducted by using Benchmarking Universal Single-Copy Orthologs (BUSCO) v3.0.2 [4]. Protein coding genes, rRNA and tRNA genes of L. taiwanensis strain K_LL004 were functionally annotated and predicted through Rapid Annotation using Subsystem Technology (RAST) v2.0 [5]. The functional categorization of all predicted Protein coding genes was performed using Clusters of Orthologous Groups (COG)-based EggNOG-mapper v2 [6]. Potential virulence factors and antibiotic resistance in L. taiwanensis strain K_LL004 were predicted using the BLASTn method according to the Virulence Factor Database (VFDB) and the Comprehensive Antibiotic Resistance Database (CARD) [7,8].
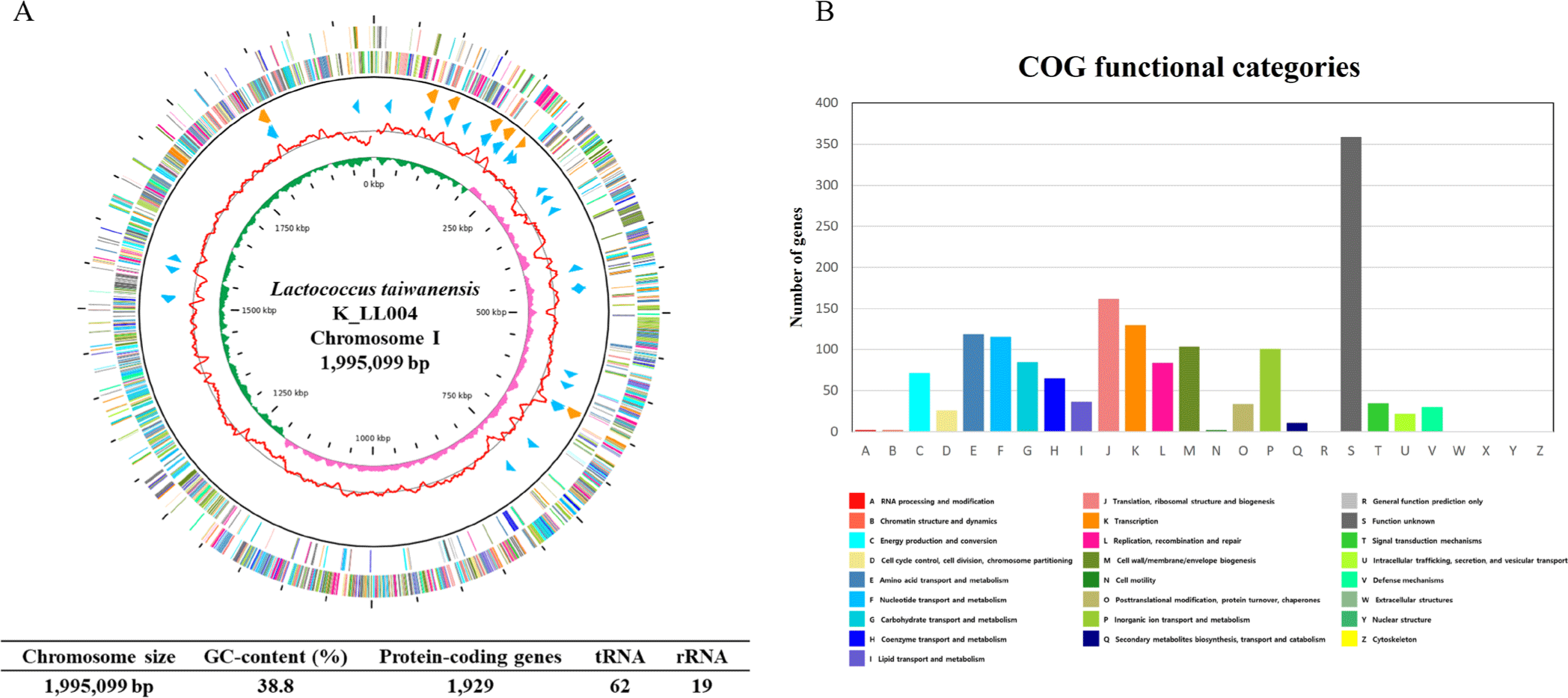
The complete genome of the L. taiwanensis strain K_LL004 contains one circular chromosome (1,995,099 bp) with a guanine + cytosine (GC) content of 38.8%, 1,929 predicted protein-coding sequence, 19 rRNA genes, and 62 tRNA genes. The genome feature and map of L. taiwanensis strain K_LL004 are illustrated in Table 1, Figs. 1A and 1B.
Table 1.
Genome features of Lactococcus taiwanensis strain K_LL004
|
Property |
Term |
|
Average genome coverage |
449x |
|
Chromosome length (bp) |
1,995,099 bp |
|
No. of contig |
1 (chromosome) |
|
Guanine + cytosine (G + C) content (%) |
38.8 |
|
Protein-coding genes |
1,929 |
|
rRNA genes |
19 |
|
tRNA genes |
62 |
|
Genbank Accession No. |
CP070872 |
Download Excel Table
Fig. 1.
Genome map of Lactococcus taiwanensis K_LL004 and the functional categorization of predicted protein coding genes.
The outer circle denotes the locations of all annotated open reading frames (ORFs), and the inner circle with the red denotes guanine + cytosine (GC) content. Pink, and green peaks denote GC skew. The orange, and sky-blue arrows denote the rRNAs, and tRNA operons, respectively. The annotated ORFs are colored differently based on the Clusters of Orthologous Groups (COG) assignments (A). COG functional categories of predicted protein coding genes (B).
Download Original Figure
It was confirmed that the L. taiwanensis strain K_LL004 has genes which encodes enzymes like beta-glucosidase (EC 3.2.1.21 BG) and beta-xylosidase (EC 3.2.1.37 xyl3), which plays an important role in beta-glycoside metabolism and xylose utilization, respectively. These enzymes are known to hydrolyze the plant cell wall polysaccharides [9]. In addition, the genome of L. taiwanensis strain K_LL004 didn’t show presence of any virulence factors and antibiotic resistant genes, indicating that L. taiwanensis strain K_LL004 can be speculated as a potential probiotic candidate with an ability to hydrolyse plant polysaccharides.
NUCLEOTIDE SEQUENCE ACCESSION NUMBERS
The complete genome sequences of Lactococcus taiwanensis K_LL004 were deposited in GeneBank under the accession numbers CP070872.1. The BioSample accession number is SAMN17981207, and BioProject accession number is PRJNA224116.