INTRODUCTION
Forage is the most economical type of feed utilized for beef and milk production. Feed costs during beef production could be reduced by efficient production and utilization of forage with high yields and high nutritional value [1]. Moreover, forage-based production systems are low-input approaches that are well suited to satisfy the demands of meat retailers and consumers for the production of beef that is ecological and animal-friendly. Hanwoo (Bos taurus coreanae), a major beef cattle raised for highly marbled beef in Korea, are reared using a combination of roughage and commercially available feeds [1,2]. The consumption of livestock products in Korea has increased, thus, the demand for roughage also increased. The local farmers opt to utilize domestically grown crops or crop silages in place of or in addition to imported forages in total mixed ration (TMR) production [2-4]. Hence, in recent years, domestic forage has been utilized to reduce the necessity for imported forage.
TMR is generally prepared by mixing concentrates and traditional roughages such as silage, forage, and hay; however, due to pasture shortages, many countries mostly rely on imported feed resources [5]. In 2020, Japan, China, United Arab Emirates, Korea, and Saudi Arabia were the major importers of high-quality forage crops primarily from the United States, Australia, Canada, Spain, and Italy [6]. Imported forage is more costly than locally grown forage due to higher transportation costs, which account for the majority of production costs [3,5,7]. As of 2020, the overall forage consumption in Korea is 4.82 million tons, wherein the domestic production is 3.92 million tons. Despite the fact that the cultivated area of forage in Korea has expanded greatly compared to the past, the domestic forage self-sufficiency rate has increased to 81.4%, 26.1% of which is accounted for high-quality forage [8]. The livestock sector’s reliance on high-quality forage drives the need for imported forages in Korea [9]. In addition, farmers tend to believe that domestically produced forages are of low quality compared to imported forages, which is not always the case [10-12]. Among imported high-quality forages, alfalfa (Medicago sativa), timothy (Phleum pratense), and oat (Avena sativa) hays are commonly used forages in Korea [10,11,13]. Imports of hay from the USA account for more than 80% of overall hay imports [13]. Alfalfa is a very important forage for livestock feeding due to its high crude protein content; however, alfalfa is rarely cultivated in Korea and the main source is imported hay. About 200,000 tons of alfalfa hay per year was imported, which accounts for 20% of the total imported hay in Korea [14,15]. However, there has been great interest in using good quality domestic forages such as Italian ryegrass (Lolium multiflorum Lam., var. italicum, IRG), whole-crop barley, and whole-crop rice as a replacement for imported forages to reduce the production cost. IRG is a highly valuable crop grown domestically as a winter forage crop due to its feed value, good nutritive quality, high yield, and palatability [2,16-18] and has the potential to reduce the necessity for imported forages in TMR production. In Korea, IRG accounts for 97% of the total cultivated area of winter forage crops and 59.9% of the total forage production in 2017. In the southern area of Korea, particularly in Jeonnam province, 62% of the production area was used for the cultivation of IRG [7,19,20]. Due to its high protein content and energy efficiency, IRG has become increasingly popular among beef producers as a roughage source. It can be provided alone (hay or silage) or as a component of TMR [2,5]. Domestically grown IRG had reduced pasture cost by up to 30%–50% compared to imported forages [19]. The replacement of imported forage with domestically grown high-quality roughage or crop silages has environmental and economic advantages, and it may reduce the feeding costs and the need for imported feed ingredients. This study was conducted to replace imported forages such as alfalfa and timothy hays by utilizing domestic Italian ryegrass silage (IRGS) in TMR production. Thus, the study evaluated the IRGS as a replacement ingredient for imported forages in TMR production and its effect on rumen fermentation, growth performance, blood metabolites, and rumen microbial communities of Hanwoo heifers during the growing period.
MATERIALS AND METHODS
The experimental protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Sunchon National University (approval number: SCNU-IACUC 2018-01). The study was carried out at the experimental farm of Sunchon National University and in the Ruminant Nutrition and Anaerobe Laboratory, Department of Animal Science and Technology, SCNU, Jeonnam, Korea. The TMR diets were formulated to replace or mix the forages (oat hay, timothy, and alfalfa hay) with IRG and corn silages. Three TMR diets were used: i) TMR containing oat hay, timothy, and alfalfa hay (CON), ii) TMR containing 19% of IRGS (dry matter [DM] basis) (L-IRGS), and iii) TMR containing 36% of IRGS (DM basis) (H-IRGS). The composition of the TMRs is shown in Table 1.
TMR diet: CON, total mixed ration containing oat hay, timothy, and alfalfa hay; L-IRGS, total mixed ration containing 19% of Italian ryegrass silage; H-IRGS, total mixed ration containing 36% of Italian ryegrass silage.
Vitamin-mineral supplement contained vit. A 2,650,000 IU, vit. D3 530,000 IU, vit. E 1,050 IU, niacin 10,000 mg, Mn 4,400 mg, Zn 4,400 mg, Fe 13,200 mg, Cu 2,200 mg, iodine 440 mg, and Co, 440 mg/kg of Grobic-DC provided from Bayer Health Care (Leverkusen, Germany).
NSC was calculated according to the formula: NSC = 100 – (NDF + CP + EE + Ash) [71].
The feeding trial was conducted for 140 days. Twenty-seven Hanwoo heifers (225.11 ± 10.57 kg, 8 months old) were used in the study. The heifers were randomly distributed into three groups of nine heifers each to evaluate three TMR diets (CON, L-IRGS, and H-IRGS), wherein 3 animals were allotted in each pen. TMR diets were prepared weekly, animals in each pen were fed once daily (08:00) for a total period of 140 days and had free access to mineral block and water. Individual pens were equipped with an electronic feed bunk monitoring system that enables monitoring of the feed intake of the animal.
The initial and final body weight (BW) of individual heifers was recorded before feeding. Average daily gain (ADG) was calculated based on the difference in BW measured in different periods divided by the number of days of the experimental period, while the feed conversion ratio (FCR) was expressed as average dry matter intake (DMI) per ADG. At the end of the experimental period, ruminal fluid and blood were obtained from each heifer. Ruminal fluid was collected using a stomach tube and the rumen sample was placed in a sterile 50 mL conical tube. Aliquot samples (1 mL) were prepared in 1.5 mL microcentrifuge tubes and stored at −80°C until analysis for rumen fermentation characteristics and metagenomics sequencing. Meanwhile, approximately 5 mL of blood was taken from the jugular vein of each heifer and transferred into a vacutainer tube (SSTTM II Advance, BD vacutainer, Becton Dickinson, Franklin Lakes, NJ, USA). The serum was separated by centrifugation at 3,134×g for 5 min at 4°C (Labogene 1248R, Labogene Co., Ltd., Daejeon, Korea). Following centrifugation, serum was carefully removed using a pipette, transferred to a 1.5 mL microcentrifuge tube, and stored at −20°C until analysis. Concentrations of aspartate aminotransferase (AST), blood urea nitrogen (BUN), albumin (ALB), total protein (TP), total cholesterol (TC), and glucose (GLU) in the serum were analyzed by IDEXX Catalyst One Chemistry Analyzer (IDEXX Laboratories, Westbrook, ME, USA).
Rumen fluid was analyzed for its fermentation parameters, including pH, ammonia-nitrogen (NH3-N), and volatile fatty acid (VFA) concentrations. The pH of the ruminal fluid was measured using a pH meter (Mettler Toledo, Greifensee, Switzerland). Aliquot rumen fluid samples were transferred in 1.5 mL microcentrifuge tubes and stored at −80°C until NH3-N and VFA analyses. The frozen rumen samples were thawed at room temperature and centrifuged at 19,326×g for 10 min at 4°C (Micro 17TR, Hanil Science Industrial Co., Ltd., Incheon, Korea). Then, the resulting supernatant was used for the NH3-N and VFA concentration analyses. Ruminal NH3-N concentration was determined by a colorimetric method as described by Chaney and Marbach [21]. The concentration of VFA in the ruminal fluid was analyzed by high-performance liquid chromatography (HPLC) on an Agilent 1200 Series HPLC System (Agilent Technologies, Santa Clara, CA, USA) equipped with a MetaCarb 87H column (300 mm × 7.8 mm) under isocratic conditions (flow rate = 0.6 mL/min of 0.0085 N sulfuric acid (H2SO4); column temperature = 35°C [22,23].
Rumen samples were thawed at room temperature and centrifuged at 19,326×g for 10 min at 4°C (Micro 17TR, Hanil Science Industrial Co., Ltd.). The supernatant was discarded, and the resulting pellet was used to extract genomic DNA using a FastDNA SPIN Kit (MP Biomedicals, Irivine, CA, USA), following the manufacturer’s protocol. The extracted DNA was sent to Macrogen, Korea, for 16S ribosomal RNA (rRNA) gene amplification and metagenomics sequencing.
The amplicon sequencing approach was performed according to the Illumina 16S Metagenomic Sequencing Library protocols [7]. Amplicon libraries targeting the V3–V4 hypervariable region of the 16S rRNA gene were generated by PCR amplification which consists of two PCR steps. The genomic DNA was amplified by using a universal primer pair with Illumina adapter overhang sequences, 16S_341F (5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3′) and 16S_805R (5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3′). The PCR mixture was conducted in a final volume of 25 μL, consisting of 2.5 μL of DNA sample (~5 ng/μL), 5 μL of each forward and reverse primer, and 12.5 μL of 2x KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA, USA). The cycling condition for the first PCR was performed as follows: initial denaturation at 95°C for 3 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The amplicons were purified using AMPure XP beads (Agencourt Bioscience, Beverly, MA, USA). After purification, 2 μL of the first PCR product was used as a template for the second PCR using a Nextera XT Index primer pair (Illumina, San Diego, CA, USA). The PCR consisted of 5 μL of sample DNA, 5 μL each of Nextera XT Index primers 1 and 2, 25 μL of 2x KAPA HiFi HotStart ReadyMix (KAPA Biosystems), and 10 μL of PCR Grade Water. The cycle condition was: initial denaturation at 95°C for 3 min, followed by 8 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The final PCR amplicon was cleaned-up using the AMPure XP beads (Agencourt Bioscience). The quantity of the final PCR amplicon was evaluated according to the qPCR Quantification Protocol Guide (KAPA Library Quantification Kit for Illumina Sequencing platforms), and library quality was assessed using TapeStation D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany). Finally, the paired-end sequencing was performed on an Illumina MiSeq platform (Illumina) using v3 reagents to generate 300 bp paired-end reads.
The paired-end sequence reads (Fastq files) were obtained using the bcls2fastq package (Illuminas). Trimmomatic v0.38 was used to filter the adaptor sequences and remove low-quality sequences from the raw sequences [24] and assembled using Fast Length Adjustment of Short Reads (FLASH 1.2.11) [25]. Low-quality sequences, ambiguous sequences, and chimera sequences from the obtained sequences were removed using CD-HIT-OTU [26]. The filtered reads were then clustered and identified as operational taxonomic unit (OTU) at 97% sequence similarity using CD-HIT-OTU [26], and chimeric sequences were identified and removed using rDnaTools (https://github.com/PacificBiosciences/rDnaTool). The taxonomy of the representative sequences from the clustered OTU was assigned using Quantitative Insights Into Microbial Ecology (QIIME Version 1) [27] from the NCBI 16S rRNA database, and the taxonomy composition from phylum to species level was generated using QIIME-UCLUST [28].
Further analysis and data visualization was performed in the MicrobiomeAnalyst tool (available at: http://www.microbiomeanalyst.ca) [29] using a BIOM formatted OTU table [12] generated in Mothur [30]. Alpha diversity of each sample was assessed using the observed OTUs, Chao1 estimator, and Shannon index. Venn diagram was generated using jvenn (http://jvenn.toulouse.inra.fr/app/index.html) to illustrate the shared and unique OTUs of rumen bacterial community in Hanwoo heifers fed different TMR [31]. The hierarchical clustering heat map was visualized using the MicrobiomeAnalyst tool using the Bray-Curtis dissimilarity test and Ward clustering algorithm [29]. Linear discriminant analysis (LDA) effect size (LefSe), which uses a non-parametric Kruskal-Wallis rank sum test and performs a LDA to evaluate the effect size of each taxon, was performed in MicrobiomeAnalyst online tool.
Data were analyzed using the general linear model (GLM) procedure in Statistical Analysis Systems (SAS) version 9.4 (SAS Institute, Cary, NC, USA). Statistical comparison of data was performed using one-way analysis of variance (ANOVA), followed by Duncan’s multiple range test (DMRT). A p-value of less than 0.05 indicates a statistically significant result.
RESULTS
The results for rumen fermentation parameters are shown in Table 2. The total VFA, propionate, butyrate, and A/P ratio showed significant differences between the three TMR diets (p < 0.05). Meanwhile, no significant differences in the rumen pH, NH3-N, and acetate concentration between the three TMR diets were observed. However, NH3-N concentration tended to be lower in TMR diets containing IRGS (L-IRGS and H-IRGS) than in CON TMR. The total VFA concentration was significantly higher in CON TMR diet (p < 0.05) than in other TMR diets. Meanwhile, heifers fed CON and H-IRGS TMR diets have a higher molar proportion (p < 0.05) of propionate in comparison to L-IRGS TMR diet. The molar proportion of butyrate in the rumen was greater (p < 0.05) when heifers were fed L-IRGS TMR diet in comparison to other TMR diets. Lower acetate-to-propionate (A/P) ratio was observed in heifers fed CON and H-IRGS TMR diets compared with L-IRGS (p < 0.05).
Treatments: CON, total mixed ration containing oat hay, timothy, and alfalfa hay; L-IRGS, total mixed ration containing 19% of Italian ryegrass silage; H-IRGS, total mixed ration containing 36% of Italian ryegrass silage.
The growth performance and blood metabolites of Hanwoo heifers fed different TMRs are presented in Tables 3 and 4. The Hanwoo heifers were fed different TMR for 140 days (May to December 2017). As shown in Table 3, the BW, ADG, DMI, and FCR of Hanwoo heifers fed with IRGS-based TMR did not differ (p > 0.05) from those heifers fed with CON TMR. These results suggest that the different TMR diets did not change the overall growth performance of the heifers. Meanwhile, the total protein, albumin, AST, glucose, and total cholesterol were not affected by the TMRs diets (p > 0.05) (Table 4). The Hanwoo heifers fed with CON TMR had the highest BUN concentration (14.50 mg/dL), while H-IRGS had the lowest BUN concentration of 12.44 mg/dL (p < 0.05).
Treatments: CON, total mixed ration containing oat hay, timothy, and alfalfa hay; L-IRGS, total mixed ration containing 19% of Italian ryegrass silage; H-IRGS, total mixed ration containing 36% of Italian ryegrass silage.
Treatments: CON, total mixed ration containing oat hay, timothy, and alfalfa hay; L-IRGS, total mixed ration containing 19% of Italian ryegrass silage; H-IRGS, total mixed ration containing 36% of Italian ryegrass silage.
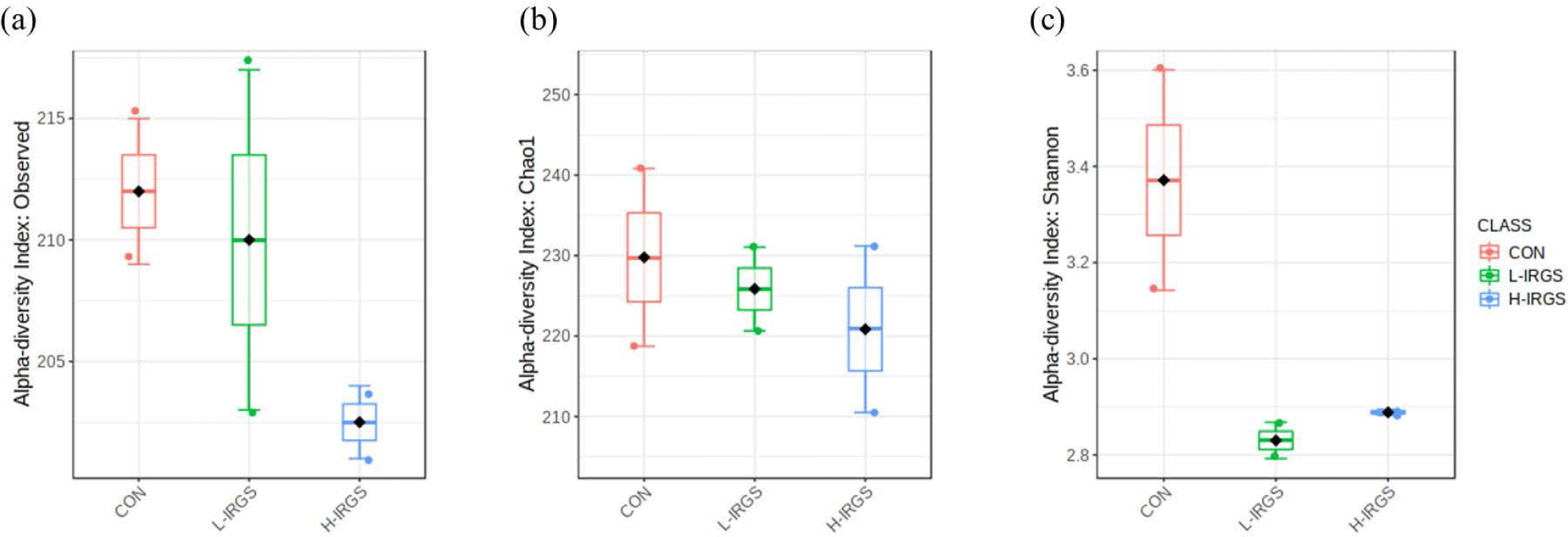
In total, 264 OTUs in the ruminal fluid were identified at 97% similarity. Observed species, Chao1, and Shannon index did not significantly vary among the rumen samples of the three Hanwoo heifer groups (p > 0.05) (Fig. 1). The number of observed OTUs and Chao1, although not significant, was higher in CON and L-IRGS than in H-IRGS (Figs. 1A and 1B). Meanwhile, the Shannon diversity index showed the most diverse species in CON compared to the L-IRGS and H-IRGS (Fig. 1C). As an indicator of microbial diversity, the Good′s coverage of all samples was more than 98% (data not shown), indicating that the obtained sequences could reflect most of the bacterial community in this study.
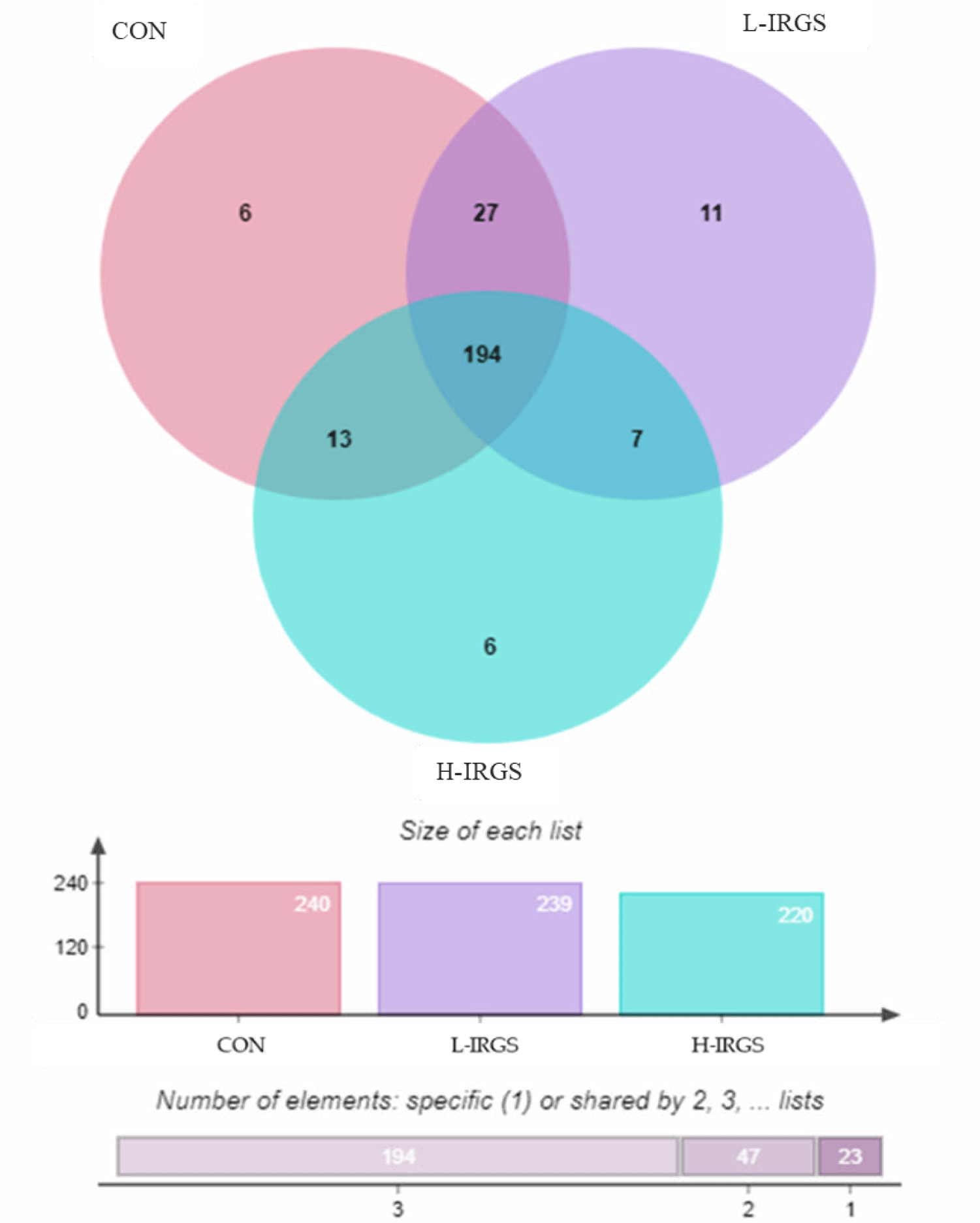
Venn diagram showed that 194 OTUs were shared by all groups (Fig. 2). For OTUs shared by two groups, CON TMR shared 27 and 13 OTUs with L-IRGS and H-IRGS, respectively, while only 7 OTUs were shared by L-IRGS and H-IRGS. Meanwhile, 6, 11, and 6 unique OTUs were observed in the CON, L-IRGS, and H-IRGS groups, respectively. The unique bacterial species found in the rumen of Hanwoo heifers fed with CON TMR were Desulfitobacterium dehalogenans, Desulfovibrio simplex, Erysipelothrix rhusiopathiae, Flavimarina pacifica, Oribacterium parvum, and Pelobium manganitolerans. On the other hand, Eubacterium hallii, Acholeplasma brassicae, Anaerosporobacter mobilis, Desulfovibrio longreachensis, Fournierella massiliensis, Fusibacter paucivorans, Lachnobacterium bovis, Merdimonas faecis, Neglecta timonensis, Prevotella enoeca, and Xanthomonas maliensis were observed only in the rumen of Hanwoo heifers fed with L-IRGS TMR. Meanwhile, Clostridium lavalense, Desulfovibrio intestinalis, Fucophilus fucoidanolyticus, Lactobacillus rogosae, Mariniradius saccharolyticus, and Robinsoniella peoriensis were observed only in the rumen of Hanwoo heifers fed with H-IRGS TMR.
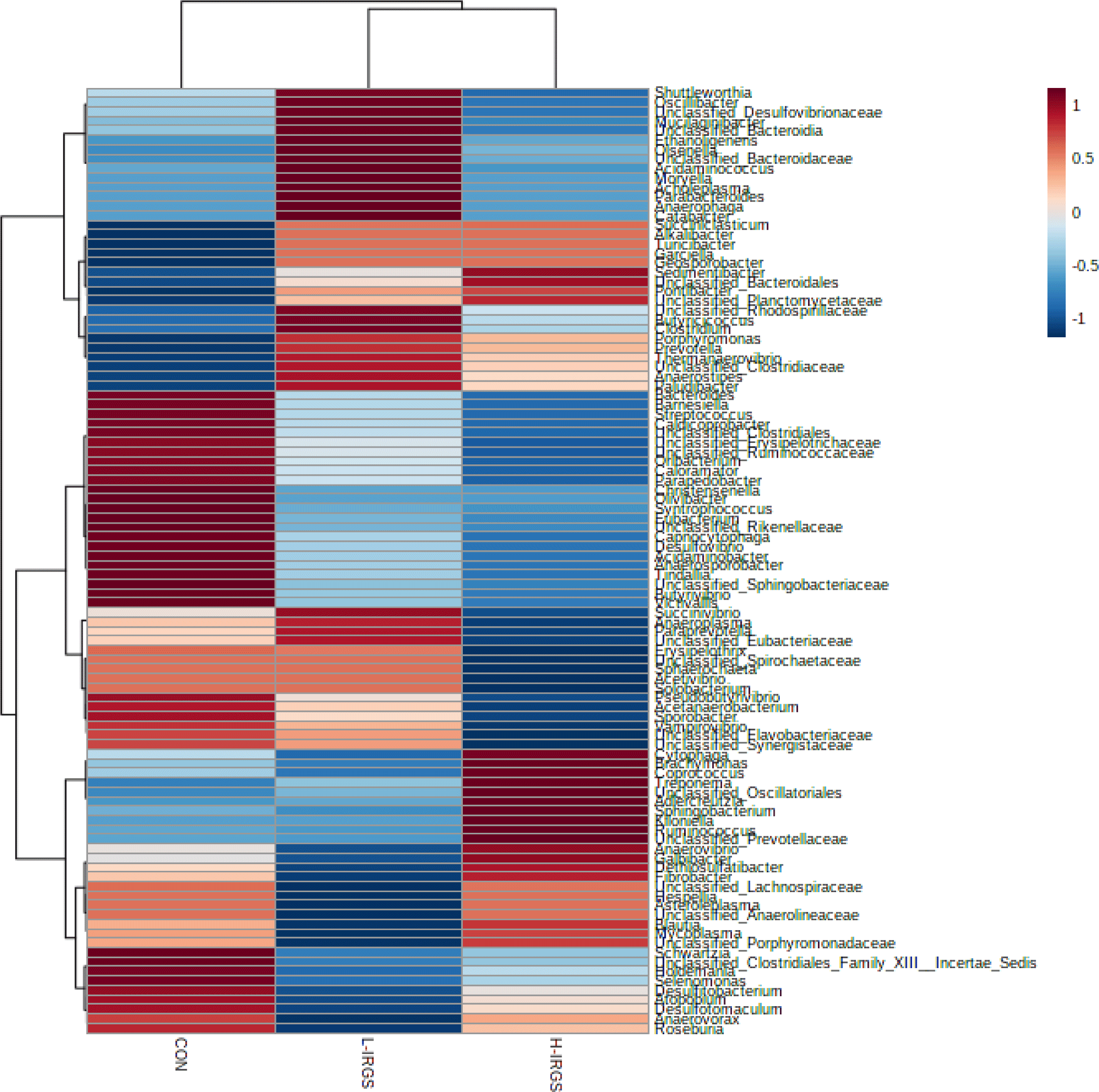
The normalized data presented in Fig. 3 shows the clustering based on the similarity of relative abundance between representative genera (row) and different TMR diets (column). The clustering in the column indicates that the bacterial community composition of the CON is different compared to the treated group (L-IRGS and H-IRGS). Meanwhile, the bacterial composition between L-IRGS and H-IRGS is relatively comparable. The genera with high and low abundance are indicated by the red and blue colors, respectively.
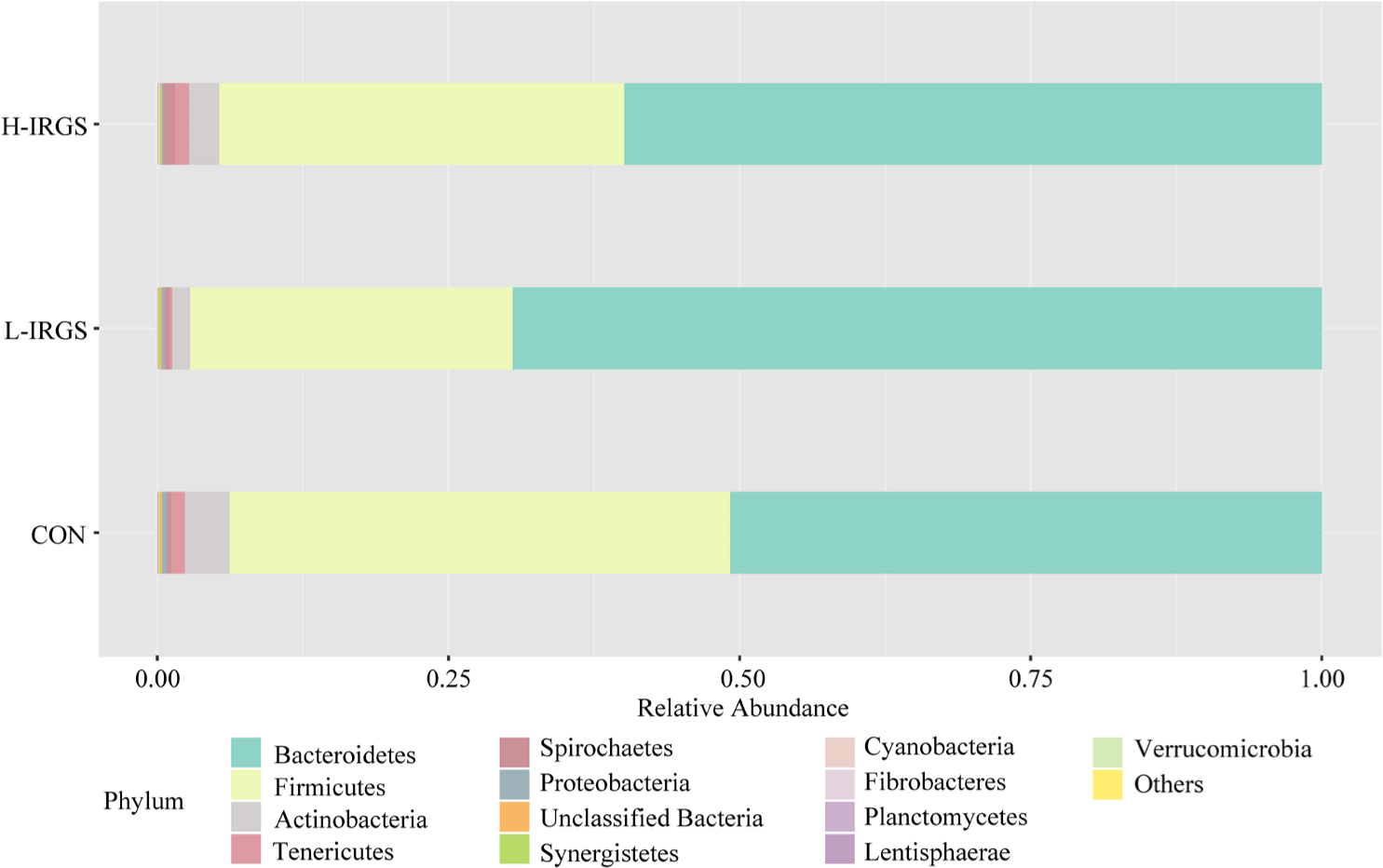
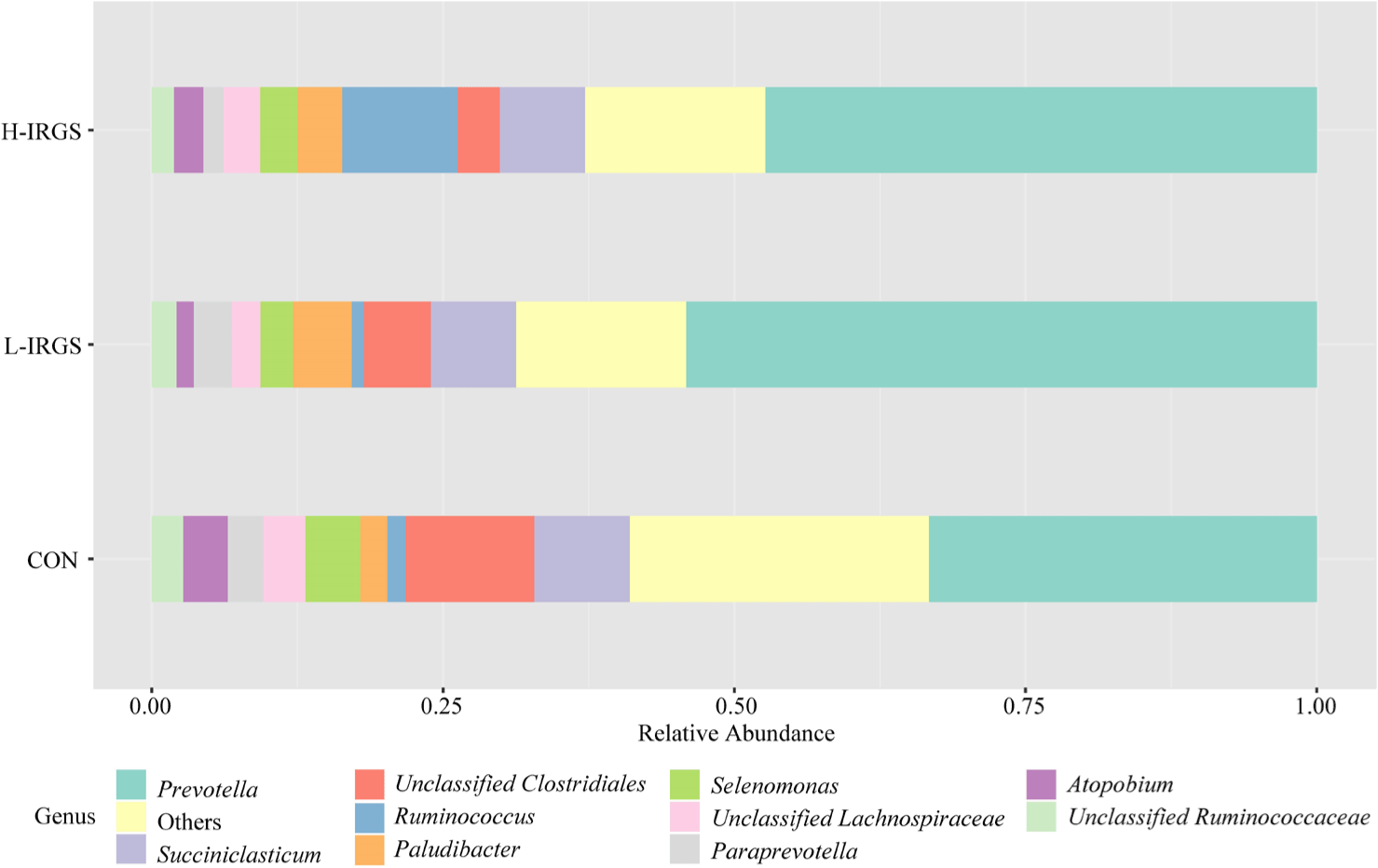
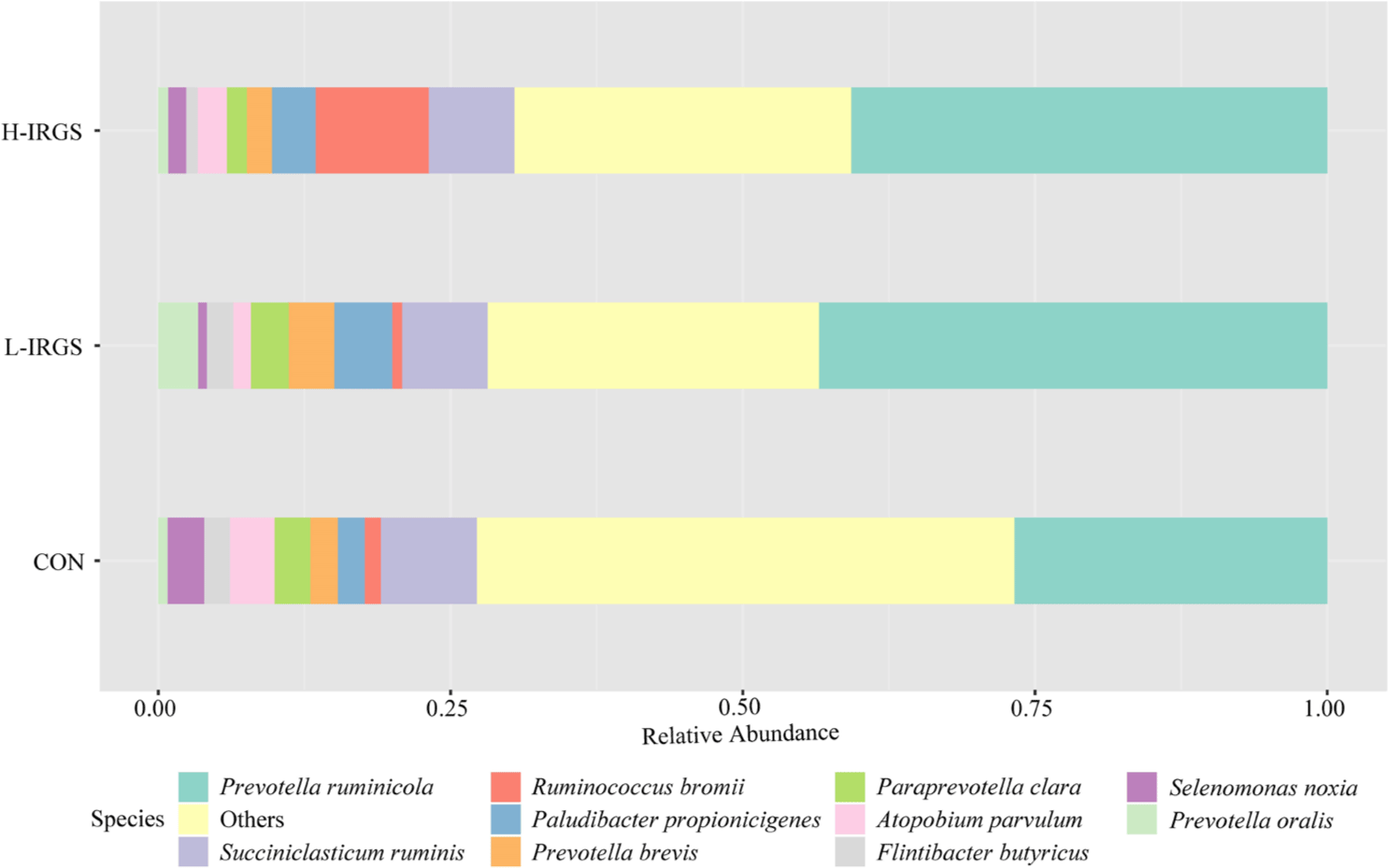
The effect of different TMR diets on the bacterial community composition is shown in Fig. 4. Illumina analysis of the bacterial community in rumen samples of the three groups of Fifteen bacterial phyla was classified in the rumen of Hanwoo heifers fed different TMR diets (Fig. 3A). Taxonomic classification showed that Bacteroidetes and Firmicutes were the predominant phyla, which accounted for 64.47% and 33.05% of the total sequences, respectively. Bacteroidetes was observed to be higher in L-IRGS-fed Hanwoo heifers accounting for 72.53% of the bacterial population. Meanwhile, Firmicutes was lower in Hanwoo heifers fed L-IRGS TMR accounting for 25.86%. At the genus level, the abundance of Prevotella increased in the rumen when Hanwoo heifers were fed with L-IRGS TMR (Fig. 5). In addition, the abundance of Ruminococcus increased in the rumen when Hanwoo heifers were fed with H-IRGS. In contrast, Unclassified Clostridiaceae and Unclassified Rikenellaceae decreased in abundance when Hanwoo heifers were fed with TMR containing IRGS. Prevotella ruminicola dominated all treatments at the species level, with relative abundances of 26.66%, 43.36%, and 40.74%, for CON TMR, L-IRGS, and H-IRGS TMR, respectively (Fig. 6). Meanwhile, the relative abundance of Ruminococcus bromii increased when the H-IRGS TMR was fed to the Hanwoo heifers.
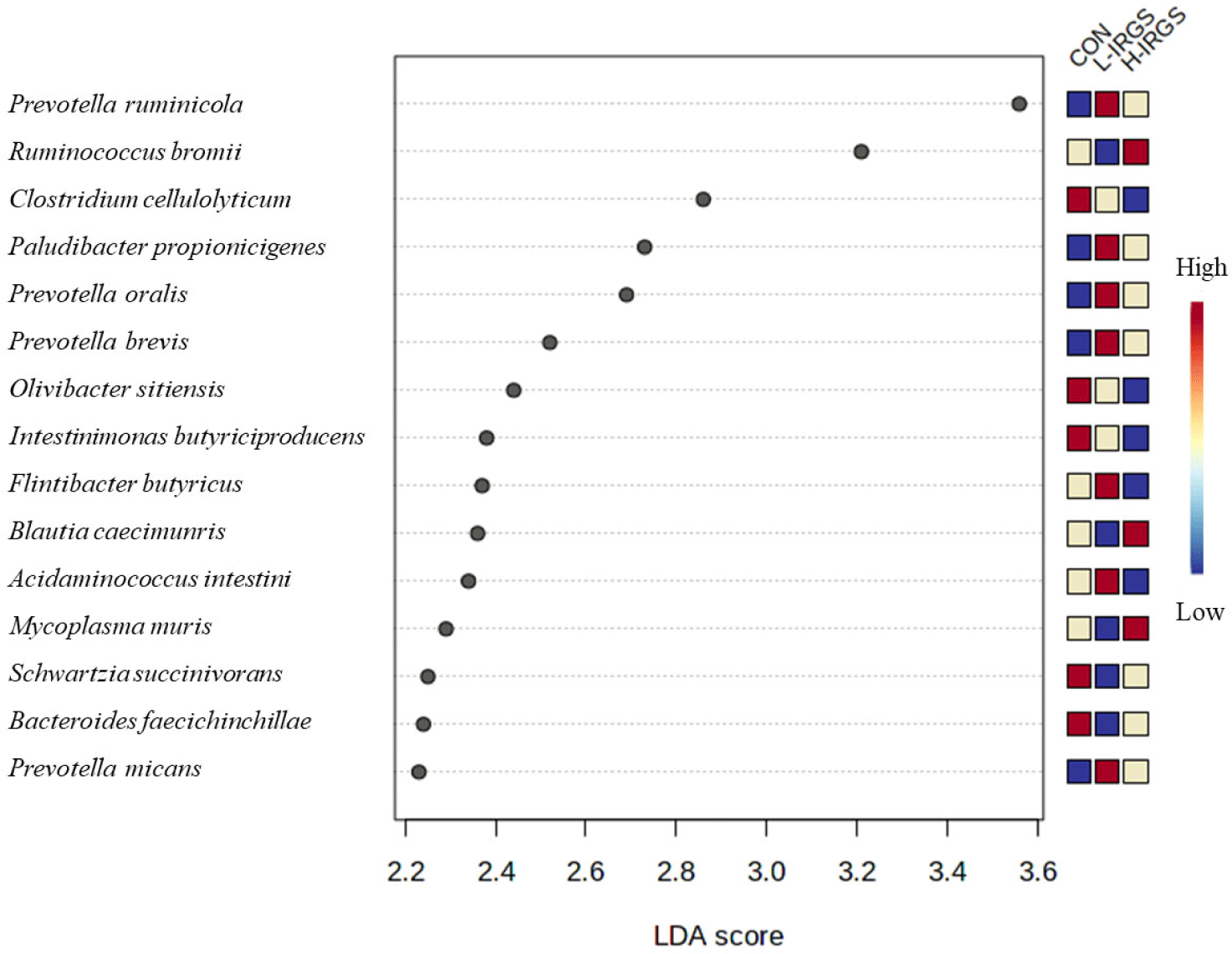
To determine the microorganisms that most likely explained significant differences among samples from the three groups (CON, L-IRGS, H-IRGS), we performed a LEfSe (Fig. 7). The LEfSe analysis revealed the top 15 significant microorganisms in the three groups, including five taxa in CON group, four taxa in H-IRGS, and six taxa in L-IRGS group. The CON group increased the abundance of Clostridium cellulolyticum, Olivibacter sitiensis, Intestinimonas butyriciproducens, Bacteroides faecichinchillae, and Christensenella timonensis. Meanwhile, bacterial species such as Ruminococcus bromii, Blautia caecimuris, Mycoplasma muris, and Galbibacter mesophilus was enriched in H-IRGS. In contrast, Prevotella ruminicola, Paludibacter propionicigenes, Prevotella oralis, Flintibacter butyricus, Acidaminococcus intestini, and Prevotella micans was increased in L-IRGS fed heifers.
DISCUSSION
This study evaluated the IRGS-based TMR as feedstuff and its effect on rumen fermentation, growth performance, and bacterial community composition in growing Hanwoo heifers. Three TMR diets (CON, L-IRGS, and H-IRGS TMR) were used in the study. In the present study, it was observed that TMR containing oat, alfalfa, and timothy hays as forages resulted in higher total concentration of VFA in the rumen than other treatments. Several studies reported that feeding TMR with the inclusion of forages such as oat, timothy, and alfalfa hays resulted in high concentration of total VFA [32-34]. In addition, the molar proportion of propionate in the rumen was significantly higher in the CON and H-IRGS TMR. Propionate is the most essential VFA precursor of glucose synthesis, and hence has a significant impact on hormonal release and tissue distribution of nutrients [35,36]. Consequently, as a result of H-IRGS TMR feeding, the propionate proportion could contribute to the improvement of nutrient supply and utilization. Moreover, ruminal propionate concentration was reported to be higher in silage feedings than in hay feedings [37]. Silages contained high value of lactic acid; hence, as lactic acid is one of the precursors of propionic acid in the rumen, the concentration of propionate is expected to increase [37,38]. Thus, the increased molar proportion of propionate in this study might be attributed to the high lactic acid concentration contained in the H-IRGS TMR. Moreover, our previous study [3] showed that lactate production was higher in TMR containing IRGS than in hay-based TMR. In addition, the increase in the molar proportion of propionate might be attributed to the rumen microbes involved in the production of propionic acid, specifically, the Ruminococcus bromii [39]. The proportion of butyrate significantly increased in the L-IRGS TMR diet, apparently due to the changes in propionate concentration in TMR diets, whereas the acetate proportion remained constant throughout the study. CON- and H-IRGS-fed heifers had higher propionate and lower acetate concentrations; thus, a lower acetate-to-propionate ratio was expected [40,41]. Acetate-to-propionate ratio across experimental diets ranges from 2.84–3.06, which is higher than the threshold of 2.78, this reflects normal ruminal fermentation [42]. Meanwhile, CON had the lowest A/P ratios compared to the other TMR diets. Russel [43] reported that ruminal fermentation end products are dependent on diet, and the A/P ratio is generally lower for cereal grains than for forages. He added that the association between the acetate-to-propionate ratio and diet has been explained by the metabolic characteristics of fiber-digesting and starch-digesting bacteria, this explanation, however, is not entirely conclusive. Some starch-digesting ruminal bacteria make large amounts of propionate, but many fiber-digesting bacteria produce large amounts of succinate, which is subsequently converted to propionate. In the present study, rumen ammonia-N concentrations did not differ among heifers fed different TMR diets; however, ammonia-N tended to be lower in TMR diets containing IRGS than in CON. The decrease in ammonia concentrations could be attributed to the increased consumption of ammonia generated by microbes with access to a readily accessible energy source, which increases microbial protein synthesis [41]. According to Satter and Slyter [44], a minimum ammonia-N concentration of 20–50 mg/L was required to sustain efficient microbial production in the rumen. In this study, ammonia-N concentration was lower than the optimal range. However, they also added that minimum levels of ammonia-N in ruminants fed fresh forage vary.
Different dietary treatments did not change the BW, ADG, DMI, and FCR of Hanwoo heifers as there was no difference between CON TMR and IRGS-based TMR diets. These results imply that the TMR diets did not adversely affect the overall growth performance of the heifers. Conversely, as the BW, ADG, DMI, and FCR were not affected, this could suggest that IRGS can be used as an alternative to high-quality imported forages in TMR production. IRGS has been utilized as feed due to its good nutritive value, high yield, good palatability, and is cheaper compared to imported high-quality forages such as alfalfa and timothy. Baldinger et al. [45] reported that the combination of IRGS and corn silage, which are both energy-rich forage, improved the DM intake. In this study, the inclusion of IRGS and corn silage in the TMR diet did not adversely change the rumen fermentation and growth performance of the heifers, implying that it is comparable to the control diet containing the imported forages. Thus, IRGS with corn silage can replace the imported forages in the TMR diet without compromising the rumen fermentation parameters and growth performance of the heifers. Furthermore, the TMR diets in this study have similar NDF values. Several studies reported that when the NDF content of a diet was the same, the forage source did not influence the ADG and growth efficiency in cattle [46-48]. Similarly, the NDF content of the TMR diets in our study was comparable, which might explain why there was no difference in the ADG and growth performance among the heifers. The BUN was highest in the CON group and lowest in the H-IRGS group. Dietary N-to-energy ratio, forage intake level, protein degradability in the rumen, as well as dietary carbohydrate amount, and liver and kidney function in ruminants can directly influence the BUN [49]. The BUN is a good indicator of rumen ammonia concentrations, closely related to the solubility of nitrogenous compounds fed in the animal [50]. In addition, it serves as an indicator of microbial protein balance and dietary protein efficiency [49,51]. Our findings showed that BUN was significantly higher in the heifers fed CON TMR. Higher concentration of BUN in heifers fed CON TMR were consistent with higher ruminal NH3-N contents, indicating lower dietary nitrogen (N) efficiency. Meanwhile, a lower concentration of BUN was observed in heifers fed H-IRGS TMR, which indicates high nitrogen metabolism capacity [52]. However, the BUN might be affected by similar starch and fermentable energy content of diets. According to DePeters and Ferguson [53], the ruminal NH3-N and BUN concentrations are highly correlated. In this study, the ruminal NH3-N was not affected by the TMR diets; however, NH3-N tended to be lower in TMR diets containing IRGS than in CON. improve N use efficiency through lower rumen NH3-N However, numerically higher rumen ammonia was observed in heifers fed TMR containing alfalfa hay. Heifers fed with IRGS-based TMR had higher DMI than heifers receiving the CON TMR diet. Moreover, the FCR of heifers receiving IRGS-based TMR was higher than those receiving CON TMR. Consequently, the growth of heifers fed IRGS-based TMR was comparable with CON TMR.
Our study explored the rumen bacterial diversity of Hanwoo heifers fed with different TMR diets. Alpha diversity indices had no significant differences between the TMR diets; however, species richness and evenness in the rumen bacterial community were higher in heifers fed with CON, which suggests that the rumen bacterial diversity of heifers fed TMR containing oat, timothy, and alfalfa hays were higher than the diversity in heifers fed TMR containing IRGS. In addition, this study observed significant shifts in the rumen bacterial populations in response to different TMR diets. Bacteroidetes and Firmicutes were found to be the most dominant phyla in the rumen of Hanwoo heifers. These findings are consistent with previous studies that reported Bacteroidetes and Firmicutes to be the most prevalent rumen phyla in ruminants [54,55]. Interestingly, the heifers fed a TMR diet containing IRGS had the highest abundance of Bacteroidetes and the lowest abundance of Firmicutes. In contrast, the CON TMR diet had the lowest abundance of Bacteroidetes (57.64%) and the highest abundance of Firmicutes. According to Hu et al. [56], high dietary energy increased the ratio of Firmicutes to Bacteroidetes and mainly increased ruminal amylolytic and propionate-producing bacteria populations. At the genus level, Prevotella is the most dominant genus found in the rumen of heifers. The most significant genera of ruminal bacteria are Prevotella, Ruminococcus, Unclassified Clostridiales, and Paludibacter were among the common genera found in the rumen of Hanwoo heifers [57]. Meanwhile, fiber-degrading bacteria such as Fibrobacter succinogenes, R. albus, and R. flavefaciens have significantly low abundance in all groups. In this study, Prevotella, which belongs to Bacteroidetes, was the dominant bacterial genus in the rumen of Hanwoo heifers. Several studies have proven that Prevotella was the most abundant in the rumen of dairy cows and beef steers and calves [58-61]. The highest Prevotella abundance was in the L-IRGS group, and the lowest abundance was in the CON TMR. Prevotella ruminicola was dominant in all TMR diet groups at the species level. In addition, the abundance of Ruminococcus was increased in the rumen when Hanwoo heifers were fed with the H-IRGS TMR diet. Ruminococcus are highly dominant in the large bowel, caecum, or rumen of many animals and humans and are one of the primary degraders of plant fiber in the rumen [62-65]. The relative abundance of Ruminococcus bromii was high in Hanwoo heifers fed with H-IRGS TMR than in other TMR diets. Prevotella spp. are amylolytic bacteria that can degrade starch, xylan, and pectin, while Ruminococcus is known as cellulose degrader in the rumen [56,66-69]. R. bromii is a specialized amylolytic bacterium that plays a significant role in utilizing and degrading resistant starch in the rumen and is associated with propionate production [56,66]. In addition, Prevotella spp. contains a membrane-bound electron transfer complex, that facilitates the reduction of fumarate to succinate, which is the substrate for the synthesis of propionate in the rumen [70]. TMR diet containing IRGS increased the relative abundance of these bacteria and may have contributed to the increased propionate production in the rumen. The inclusion of IRGS in the TMR diet increased the abundance of Bacteroidetes and mainly increased propionate-producing bacteria populations. The present study provided information on the effects of IRGS as an alternative forage source in TMR production and feedstuff for Hanwoo heifers during the growth period. Hanwoo heifers fed a TMR diet containing IRGS at 36% DM increased the molar proportion of propionate and increased the abundance of R. bromii in the rumen. This could suggest that the inclusion of a high amount of IRGS in TMR increased the abundance of propionate-producing bacteria, as well as the propionate production to enhance the energy harvest in the rumen. In addition, the effect of IRGS-based TMR on the growth performance of heifers is similar to those heifers fed CON TMR diet. Therefore, IRGS may be used as an alternative ingredient for imported forages in TMR production. The H-IRGS TMR decreased the total VFA production, reduced the acetate-to-propionate ratio, and improve N use efficiency through lower rumen NH3-N.
