INTRODUCTION
Influenza A virus (FLUAV) is a representative RNA virus that can be transmitted from wild birds to poultry and can spread throughout the body and respiratory tract, causing serious diseases [1,2]. Highly pathogenic avian influenza (HPAI) viruses cause serious infections in poultry and humans [3,4], and if pathways for RNA viruses to circulate and produce modified copies by mutation, genetic recombination, and genetic reassortment were to develop, a global pandemic could occur, causing significant economic losses [2,5,6]. Although there is a focus on developing poultry vaccines, avian influenza (AI) poses challenges because mutations are common and the vaccine’s efficacy is reduced in immunocompromised individuals. This has fueled debate over vaccine effectiveness among scientists, as poultry HPAI outbreaks have been reported in countries where vaccinations have already been administered [7–10].
Animal host cells have proteic mechanisms that suppress viruses [11]. Representative proteins primarily responsible for cellular immunity include protein kinase R (PKR), 2’,5’-oligoadenylate synthetase (OAS)/ribonuclease L, adenosine deaminase RNA specific, and myxovirus (influenza virus) resistance 1, interferon (IFN)-inducible protein p78 (mouse) (Mx1) [12,13]. These proteins are reported to be expressed within cells after virus or IFN stimulation, conferring infection resistance [14–17]. However, their specific immune defense mechanisms remain unclear, and although we can now artificially synthesize the AI virus, its ability to transform in vivo makes interpreting experimental results difficult [10,13,18]. Known candidate genes related to AI resistance include Mx1, TLR7, RIG-I, MDA-5, NLGN4, and GAP43 [19–23], but no single gene has produced clear, consistent results [23,24]. This is because there are specific regions that exhibit antiviral effects against specific viruses, and different genotypes exist, each with distinct single nucleotide polymorphisms (SNPs) [19,25,26]. The existence of highly resistant genotypes underscores the need for active research to elucidate their genetic specificity.
Due to the limitations of poultry vaccines, there is an urgent need to identify AI-resistant genes and their relevant SNPs, as they can be used to build a biological defense system by replacing susceptible varieties with resistant ones. As the ultimate goal of this research is to prevent the spread of AI from wild birds to chickens, we selected breeds raised in Jeju, a Korean island where controlling anthropogenic risks is quite easy and AI incidence is relatively low (Fig. 1). After infecting chicken embryo fibroblasts (CEFs) from these breeds with three types of AI viruses, we performed gene expression profiling, identified SNPs through RNA-sequencing (RNA-seq), and confirmed the phenotypes of the identified genes in vitro to provide an appropriate strategy for selecting and developing AI-resistant chickens.
MATERIALS AND METHODS
We used two HPAI strains, H5N1 and H5N6, and one low pathogenicity avian influenza (LPAI) strain, H9N2. Strain H5N1 was obtained from the Korea Veterinary Culture Collection (KVCC-VR1300046). We isolated H5N6 from duck (Anas platyrhynchos) feces and H9N2 from laying hens (Gallus gallus). Experiments employing HPAI were approved by the Institutional Biosafety Committee of Jeonbuk National University (approval no. JBNU 2021-06-005) and performed in accordance with current guidelines and regulations for high-risk pathogen handling.
A total of 70 chickens were purchased from seven Jeju Island farms (Fig. 2A), with age and sex being random. This work was approved by the Institutional Animal Care and Use Committee of the Veterinary Research Institute, Jeju Special Self-Governing Province (approval No. 2020-2) and performed in accordance with current animal testing regulations. We anesthetized the animals by intravenous injection with a combination of ketamine and xylazine (20 and 2 mg/kg, respectively), and after confirming the absence of the flexion reflex, chickens were euthanized by intracardiac injection with T-61 (0.3 mL/kg).
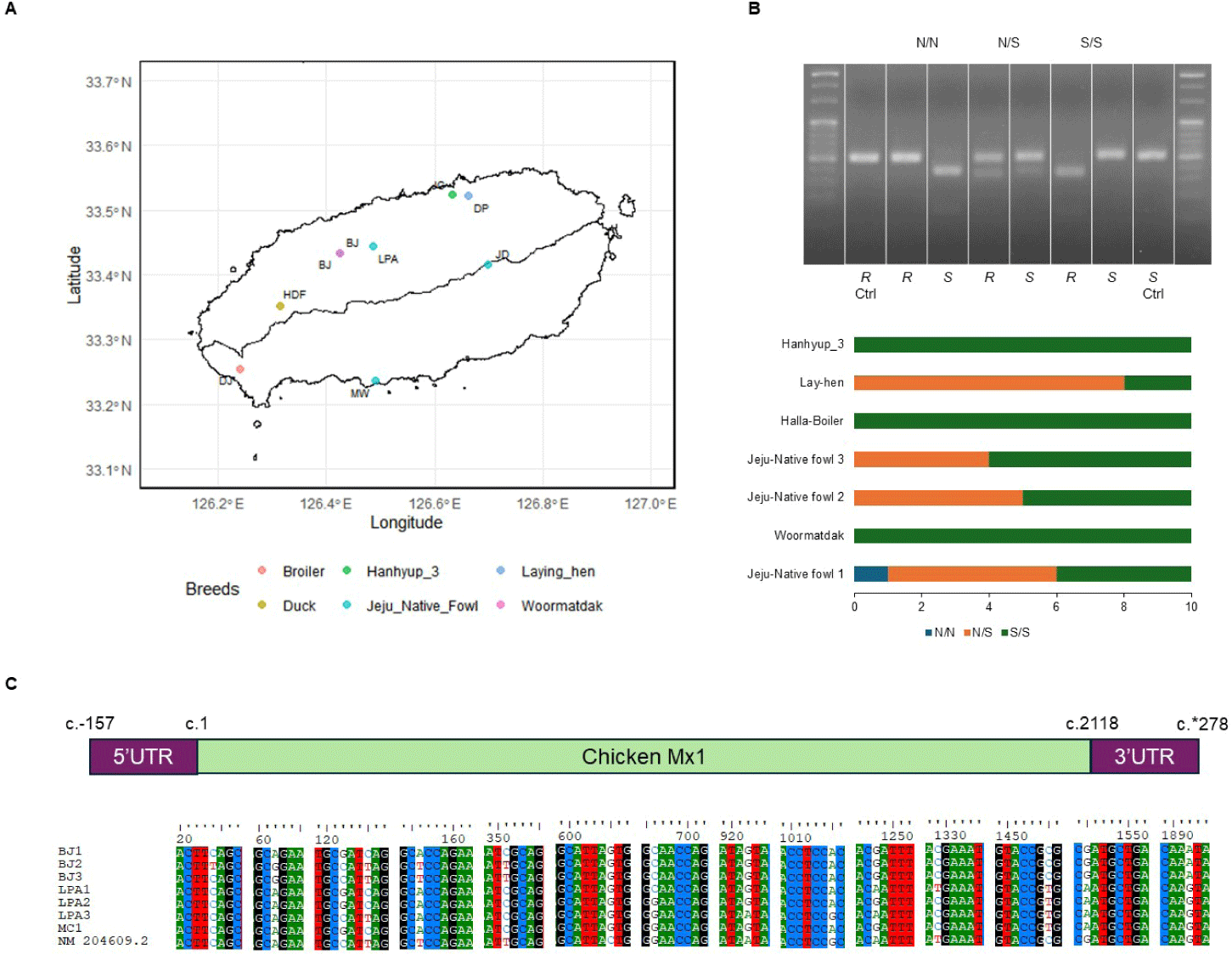
Eggs from Jeju Native chicken and Woormatdak breeds were sourced from BJ Farm. Embryonic tissues from the fertilized eggs of Jeju Native chickens, incubated for 11 d, were used for DNA extraction and CEF isolation. Samples were categorized into three groups based on their Mx1 genotype at position 631, i.e., serine (S) vs. asparagine (N): AN (Jeju Native chicken 1, Mx N/N type), AS (Jeju Native chicken 1, Mx S/S type), and MS (Woormatdak, Mx S/S type). Primary CEFs were cultured from these embryonic tissues [27], and cells between passages 5 and 8 were used for subsequent experiments (Table 1 and Fig. 3).
| Group | Breed | Mx1 genotype | FLUAV | Biological replicate |
|---|---|---|---|---|
| Control | A | N | N | 4 |
| A | S | N | 4 | |
| M | S | N | 4 | |
| Infection | A | N | H5N1 | 4 |
| A | S | H5N1 | 3 | |
| M | S | H5N1 | 4 | |
| A | N | H5N6 | 4 | |
| A | S | H5N6 | 3 | |
| M | S | H5N6 | 4 | |
| A | N | H9N2 | 4 | |
| A | S | H9N2 | 4 | |
| M | S | H9N2 | 4 |
Genome-based polymerase chain reaction- restriction fragment length polymorphism genotyping
DNeasy Blood & Tissue kits (Qiagen, Darmstadt, Germany) were used to extract DNA from the limbs of chick embryos. A PCR-RFLP analysis targeting the Mx variation at position 631 was then conducted for CEF classification. The PCR products produced by primers targeting the S type and N type (Table 2) were digested with RsaI (ER1122, Thermo Scientific™, Vilnius, Lithuania) and SspI (ER0772, Thermo Scientific™), respectively.
CEFs were divided into three groups and inoculated with H5N1 (n = 11), H5N6 (n = 11), and H9N2 (n = 12) (multiplicity of infection = 0.1). An uninfected control group of CEFs (n = 12) was also included. After 7 h, total RNA was harvested from each CEF strain using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The RNA samples were submitted to Macrogen (Seoul, Korea) for RNA-seq. AI infection was confirmed by observing cytopathic effects (CPEs) using crystal violet staining, with AI resistance quantified as the 50% tissue culture infective dose (TCID50) [28,29].
Libraries were prepared using TruSeq Stranded mRNA LT Sample Prep kits following the TruSeq Stranded mRNA Sample Preparation Guide (Part # 15031047 Rev. E or # 1000000040498v00), and 151 bp paired-end sequencing was performed on the Illumina platform. For bioinformatic processing, we used FastQC (v0.11.7) to ensure the RNA-seq data quality, and adaptor sequences, contamination, and low-quality reads were removed using Trimmomatic (v0.38). Reads were then mapped using HISAT2 (v2.1.0) through the Bowtie2 aligner (v2.3.4.1). For each sample, we quantified all known mRNA transcripts in the Ensembl chicken gene annotation (GRCg6a) using StringTie (v2.1.3b). Differentially expressed genes (DEGs) were identified as those exhibiting a |fold change (fc)| ≥ 2 and raw p-value < 0.05 using the R packages DESeq2, when the number of biological replicates per group was ≥ 3, or edgeR, when it was < 3 [30].
To identify SNPs across various genes, functional genomics reads from each available replicate of the CEFs were aligned to the reference genome using STAR (v2.6.0c) in two-pass mode to improve alignment accuracy. Gene annotations in GTF format were incorporated, and the alignments were output as coordinate-sorted BAM files. These BAM files were indexed using SAMtools (v1.9) for efficient data processing. SNP detection was performed using BCFtools (v1.9). Variants were called by aggregating sequencing data at each genomic position with bcftools mpileup, followed by identification using bcftools call. To ensure the reliability of the detected variants, the resulting VCF files were filtered with the vcfutils.pl varFilter script included in BCFtools. Variants meeting stringent criteria—a minimum quality score of 20 and a minimum read depth of 100—were retained to ensure high confidence and sufficient coverage. Special attention was given to SNPs within target genes for subsequent analyses.
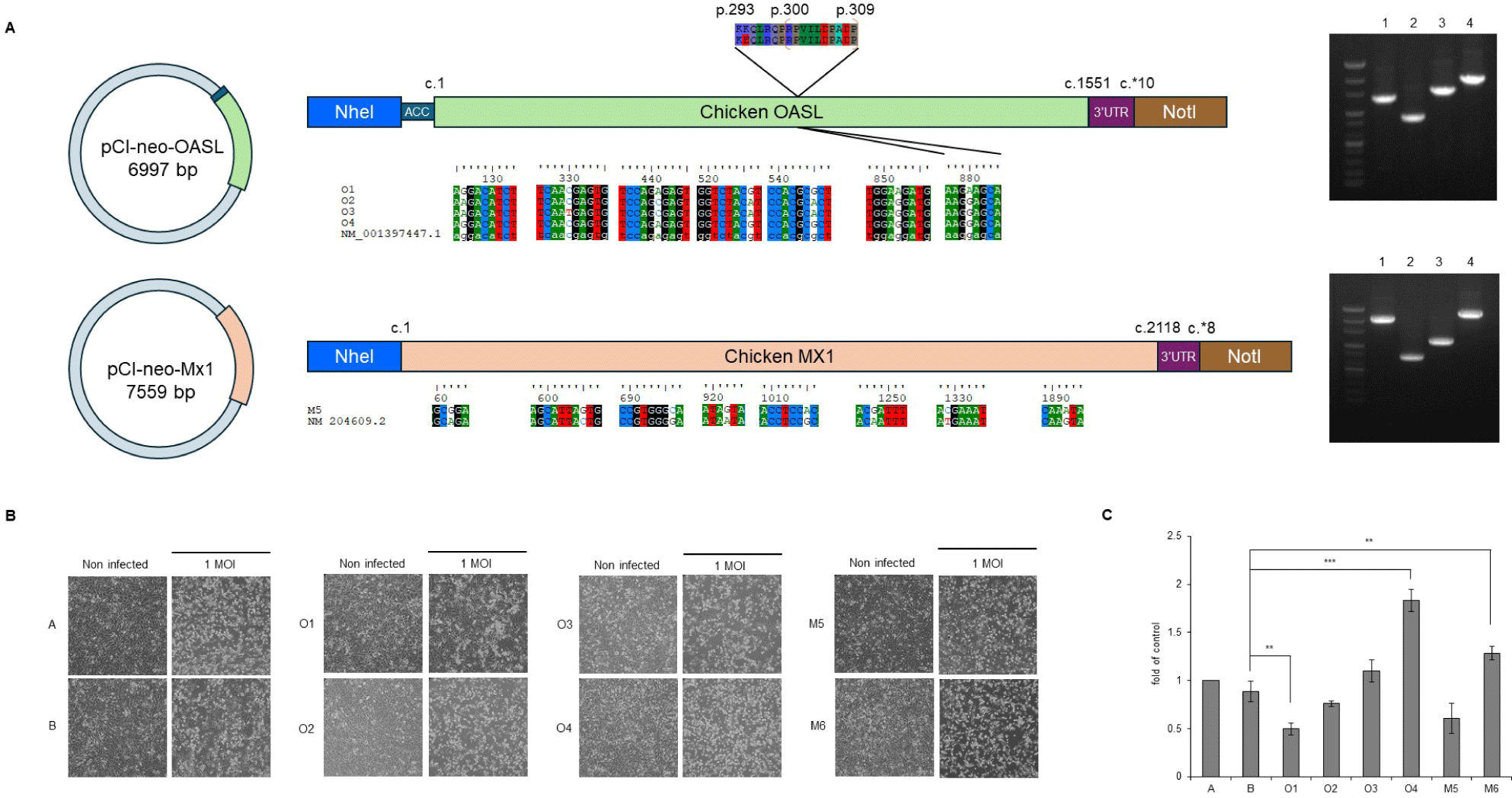
To construct the target genes, RNA from the CEFs and RNA to cDNA EcoDry Premix (Double Primed) (Clontech Laboratories, Kusatsu, Japan) were used for cDNA synthesis, and then cDNA from the Mx1 and OAS-like (OASL) mRNA fragments (identified based on accession nos. NM_204609.2 and NM_001397447.1, respectively) were amplified with primer pairs targeting the full-length sequences (Table 2) using SeqAmp™ DNA Polymerase (Clontech Laboratories). The purified amplicons were cloned into pTOPcloner™ Blunt (Enzynomics, Daejeon, Korea). Each clone was then cloned into the expression vector pCI-neo (Promega, Fitchburg, WI, USA), creating plasmids pCI-neo-Mx1 and pCI-neo-OASL.
To assess the effects of target gene overexpression, mouse fibroblasts (BALB/3T3 clone A31) were transfected with an empty vector (control), one of two Mx1 variants, or one of four OASL variants in 100 mm dishes using FuGENE 6 (Promega). Colonies of G418 (600 µg/mL)-resistant cells were selected, and transfection efficiency was confirmed with PCR and sequencing.
RNA was isolated from Balb/3T3 cells using TRIzol Reagent (Invitrogen) and from culture supernatants using Trizol LS (Invitrogen). Quantitative PCR (qPCR) was performed using GDX Probe 1-step RT-PCR Master Mix (GDX, Seoul, Korea) on a LightCycler 2.0 instrument (Roche, Basel, Switzerland) using primers detailed in Table 2.
The resistance to H5N6 infection was assessed using TCID50 values, presented as the mean ± SD of two technical replicates from four biological replicates. Virus production by SNPs was evaluated based on the relative expression of the M gene, showed as the mean ± SD of two technical replicates from three biological replicates. Statistical significance was determined using one-way ANOVA with Tukey’s post hoc test in R.
RESULTS
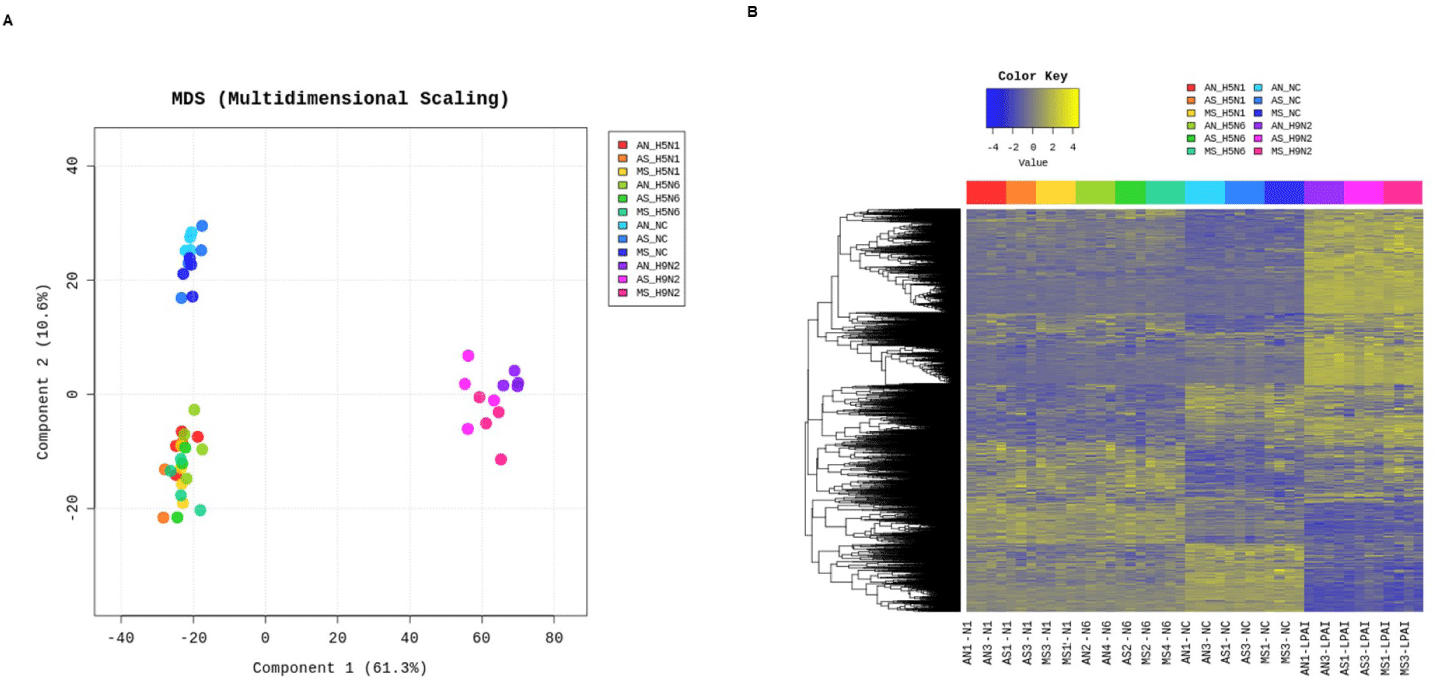
The Mx1 genotypes were confirmed for seven chicken breeds from six farms, with 10 muscle samples collected per farm (Figs. 2A and 2B). Three breeds, Woormatdak, Hanhyup, and Broiler, carried only the S/S genotype, and two, Lay Hens and Jeju Native, exhibited both N/S and S/S genotypes. Notably, while Jeju Native chickens from three farms were studied, only those raised at the BJ Farm carried all three genotypes, N/N, N/S, and S/S. Therefore, Jeju Native chickens from the BJ and Livestock Promotion Agency (LPA) farms as well as Woormatdak chickens from the BJ Farm were selected for full Mx1 cDNA sequencing using fertilized eggs. Although the Mx1 gene sequences were consistent with the reference sequence over much of their length, variations were observed. Additionally, shared sequences were identified between Jeju Native chickens from the LPA and Woormatdak chickens from the BJ Farm (Fig. 2C). Thus, CEFs representing three types, all from chickens reared separately on the same farm, the BJ Farm, were selected for further analysis: Jeju Native CEFs carrying the AI-resistant N/N Mx1 genotype (labeled AN), Jeju Native CEFs carrying the AI-sensitive S/S Mx1 genotype (labeled AS), and Woormatdak CEFs carrying the AI-sensitive S/S Mx1 genotype (labeled MS). A total of 46 samples derived from 12 eggs were included in the study. After stratifying and grouping the genotypes, each group was infected with one of the three FLUAVs (Table 1 and Fig. 3). Expression patterns formed three well-distinguished clusters (Fig. 4A): one shared by the CEFs infected with two HPAI viruses and two unique clusters corresponding to the LPAI-infected and non-infected CEFs (Fig. 4B). Gene ontology (GO) analysis revealed that, compared to the controls, the genes dysregulated in FLUAV-infected CEFs were significantly associated with GO biological process (GOBP) terms related to infection pathology, including “translation,” “peptide biosynthetic process,” “ribosome biogenesis,” and “nucleoside triphosphate metabolic process.” Additionally, many ribosomal protein genes, EIF3J, and MCTS1 were upregulated, while BTG2 and EIF2AK3 were downregulated (Fig. 5A). Notably, there were large differences between expression change patterns induced by HPAI and LPAI viruses. Under HPAI infection, GOBP terms related to immune response, inflammation regulation, and tissue repair—such as “cytokine production,” “negative regulation of cell population proliferation,” and “positive regulation of cell migration”—were significant, and genes such as FGF2, LYN, and FLT4 were upregulated, while genes such as HGF, ANGPT1, and ROR2 were downregulated (Figs. 5B, 5C, and 5D). Conversely, under LPAI infection, upregulated genes included PARK7, RACK1, and DTX3L, and downregulated genes included SIRT1, LRRK2, and WAC, all of which are associated with GOBP terms related to gene expression and protein function regulation, including “chromatin organization,” “histone modification,” and “protein modification by small protein conjugation.” Contrary to expectations, the Mx1 gene was not differentially expressed in any group (Fig. 5E).
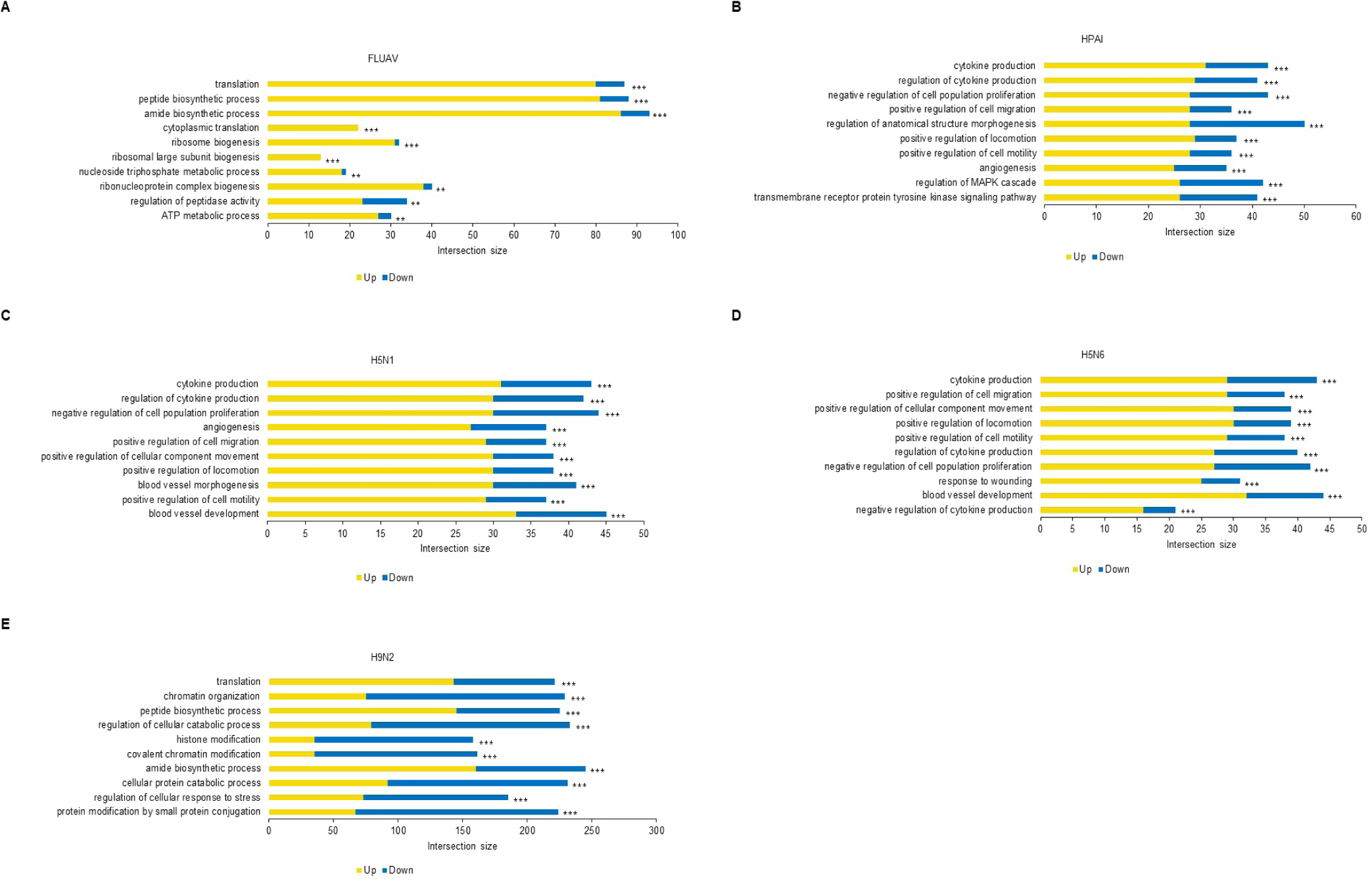
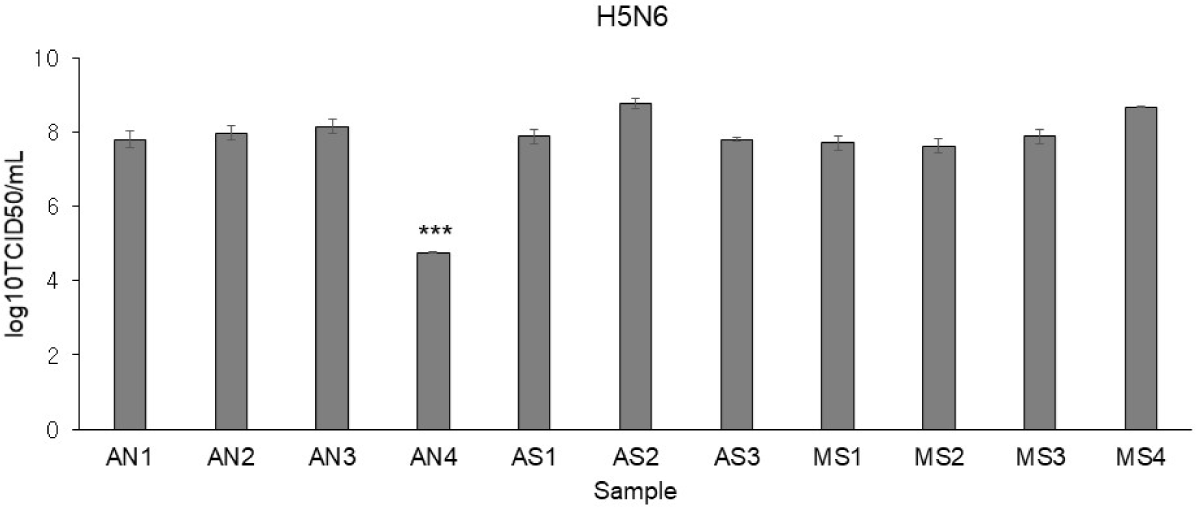
Using H5N6, virus infectivity in CEFs was confirmed by observing CPE. While it is known for conferring resistance, the AI-resistant Mx1 gene SNP could not predict H5N6 resistance alone. Interestingly, only one CEF showed resistance to H5N6 (Fig. 6). This CEF, dubbed AN4, had the Mx1 631 R/R genotype. Therefore, RNA-seq data were reanalyzed across three comparisons using 11 samples: (1) the H5N6-infected AN4 individual vs. the uninfected control AN4 individual, (2) H5N6-infected individuals AN1–AN3 vs. the H5N6-infected AN4 individual, and (3) the 10 H5N6-infected individuals (excluding the H5N6-infected AN4 individual) vs. the H5N6-infected AN4 individual. In comparison (1), 2,009 DEGs were identified in the AN4 individual depending on infection status (Fig. 7A). Notably, while the top GO term for all CEFs under H5N6 infection was “cytokine production,” it shifted to “response to growth factor” when focusing on the AN4 individuals (Fig. 7C). However, it is important to note that these terms remain closely interconnected in biological processes like inflammation, tissue repair, and immune response (Figs. 5B, 5C, 5D, and 7C). In comparison (2), 545 DEGs were identified when comparing the three infected AN1–AN3 individuals, which had the same Mx1 genotype and were from the same farm, with the H5N6-infected AN4 individual (Fig. 7A). Genes that were significantly related to immune response, including FGF10 (upregulated), NFIB (upregulated), EPHA4 (upregulated), F2RL1 (downregulated), STK39 (downregulated), and COLEC10 (downregulated), were associated with GOBP terms such as “positive regulation of phagocytosis,” “response to vitamin,” and “phosphatidylserine metabolic process” (Fig. 7D). In comparison (3), 498 DEGs were detected in all H5N6-infected CEFs relative to the H5N6-infected AN4 individual, including FGF10 (upregulated), NFIB (upregulated), ROBO1 (upregulated), GPER1 (downregulated), TGFB3 (downregulated), and LAMA1 (downregulated), which were associated with GOBP terms such as “negative regulation of cell population proliferation,” “metal ion transport,” “head development,” and “vasculature development,” indicating their potential roles in regulating proliferation, ion homeostasis, and developmental processes (Figs. 7A and 7E).
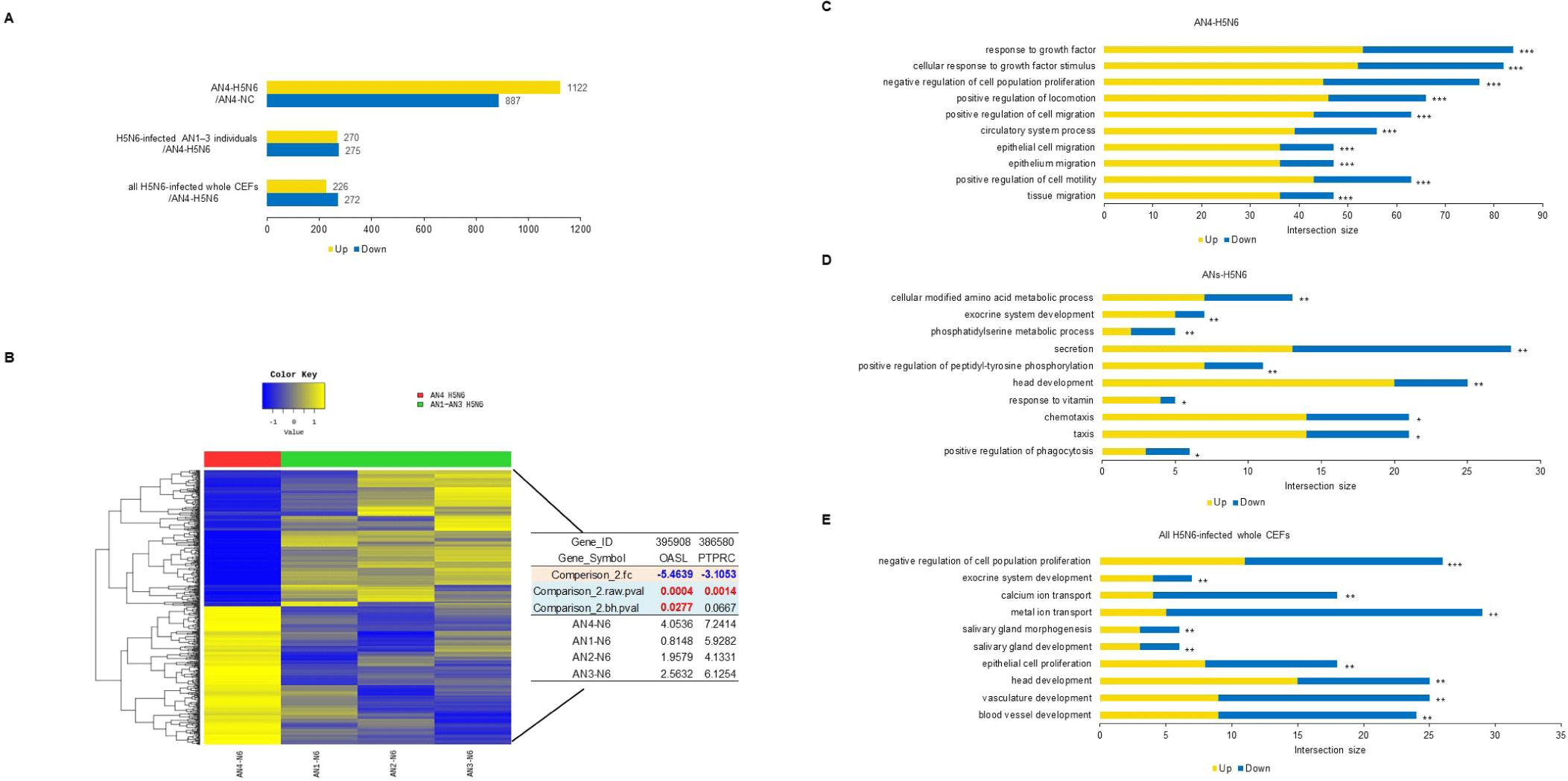
We identified GOBP terms based on specific keywords, focusing on the keyword “virus,” and investigated the associated genes. The genes identified in comparison (3) we investigated were OASL and protein tyrosine phosphatase receptor type C (PTPRC). Among the H5N6-infected AN individuals, OASL was strongly expressed in AN4 (Fig. 7B). We focused on this and performed additional single-nucleotide variant calling to determine whether OASL has an SNP associated with infection resistance, revealing a unique SNP of OASL that was only observed in the called sequence of AN4: c.G880A (p.E294K) (Fig. 8A). The full-length sequence of AN4’s OASL mRNA was extracted from AN4’s cDNA, and a construct was created enabling its overexpression. Most cells remained viable (over 80% viability), so Mx1 and OASL overexpression did not cause cytotoxicity. Thus, H5N6 virus infectivity was evaluated by observing CPE and M gene PCR. All cells exhibited morphological changes after H5N6 infection. However, cells transfected with OASL containing the c.G880A SNP showed minimal morphological changes and had the lowest expression value of the M gene (Figs. 8B and 8C).
DISCUSSION
In order to discover chicken breeds resistant to AI, which threatens public health and causes huge losses to farms every year, we performed an RNA-seq analysis by infecting fibroblasts from chickens maintained on Jeju Island with H5N1, H5N6, and H9N2, viruses mainly observed in the wild. Jeju is the optimal region in Korea for discovering and maintaining AI-resistant genes or host individuals [31]. To precisely identify host susceptibility/resistance-related genes and SNPs, DEGs were characterized, and AI infection was associated with GOBP terms related to infection pathology in the host and significantly increased the expression of PTPRC and OASL in particular (Fig. 7B). The protein encoded by PTPRC, commonly referred to as CD45, belongs to the protein tyrosine phosphatase (PTP) family. PTPs function as key signaling molecules, regulating diverse cellular processes such as cell growth, differentiation, mitosis, and oncogenic transformation [32]. Interestingly, different viruses have evolved distinct mechanisms to target PTPRC. For instance, in studies involving human immunodeficiency virus 1, PTPRC expression was upregulated during infection acquisition, potentially serving as a hallmark of increased virus acquisition risk alongside activation of the IFN response pathway [33]. In contrast, PTPRC is downregulated in T cells infected with roseoloviruses, while human cytomegalovirus and murine cytomegalovirus interfere with PTPRC signaling and transport [33]. Our findings align with observations in which PTPRC expression is upregulated upon infection acquisition, highlighting the need for a deeper understanding of the host immune response to specific viral infections.
While highly pathogenic H5N1 has attracted global attention due to its wider geographical range and the large human population it affects, another highly pathogenic virus, H5N6, deserves as much attention, if not more, due to its broad genetic range and the increasing human populations afflicted in the regions where it is found, particularly in East Asia [34]. Recently, H5N1 and H5N6 co-infections with a variety of strains [35] and reassortants [36] have been confirmed. Because H9N2 shows low pathogenicity, it has continued to spread in wild populations and has served as a donor virus contributing internal genes to newly emerging AI viruses, expanding their host range and increasing their pathogenicity and transmissibility in mammals [37]. The severity and transmissibility of diseases are determined by not only viral mutations but also host factors and/or host–pathogen interactions [38]. While LPAI infections are generally mild, they may increase the risk of secondary bacterial infections, and at least some LPAI viruses can cause systemic and fatal diseases in turkeys [39]. Therefore, it is essential to evaluate the resistance to LPAIs and HPAIs exhibited by transformants of innate immunity genes before attempting preventive measures including vaccine development. In this study, we failed to demonstrate an antiviral effect of Mx1 N631 in the CEFs using the three FLUAVs. The Mx1 gene did not show significant changes pattern in expression across the AI-infected and the uninfected control groups. This observation can be understood within several contexts. In a previous study, we characterized the diversity of PKR and Mx protein sequences in chicken breeds maintained in Japan, discovering among them an Mx gene subtype that conferred resistance to HPAI (H5N1) [19,40]. After Mx1 transfection, cells exhibited sensitivity to H5N1 when amino acid 631 in the Mx protein was S and resistance when it was N. Ewald et al. [41] evaluated whether Mx1 631 heterozygosity influences the pathogenesis of HPAI using N/N, S/S, and N/S commercial broiler strains. When challenged intranasally with an HPAI strain (H5N2), chickens homozygous for N631 were significantly more resistant than those homozygous for S631, providing evidence for an in vivo antiviral effect of Mx1. However, Schusser et al. [42], who performed infection experiments using chicken embryonic fibroblasts with Mx 631 polymorphisms and introduced reverse-transcribed viral vectors for the expression of Mx isoforms into chicken cells and embryonated eggs, found that neither N631 nor S631 isoforms provided antiviral protection against several LPAI and HPAI strains. Furthermore, they provided evidence that the antiviral effect of type I IFN in chicken cells is independent of Mx using short interfering RNA-mediated knockdown, suggesting that other IFN-inducible factors must contribute to FLUAV suppression. Considering such opposing results, we suggest that it is unreliable to predict HPAI resistance based on Mx1 expression level alone without a close examination of multiple key factors. Instead, the presence of specific SNPs, along with interactions between other immune-related genes, may play a more critical role. Lee et al. [43] analyzed miRNA expression profiles following HPAI H5N1 virus infection in resistant and susceptible Ri chicken lines. Chickens were classified based on A/G alleles of specific genes, including Mx. The study identified potential regulation of miRNAs targeting candidate genes associated with immune responses to HPAI infection. This highlights the importance of shifting from single-gene analyses to exploring broader genetic networks contributing to resistance.
We confirmed that a chicken breed with both the AI-resistant Mx1 (p.S631N) and Jeju-specific OASL subtype (p.E294K) exhibited HPAI (H5N6) resistance (Figs. 2C, 6, and 8). The OASL family of antiviral proteins plays a crucial role in the antiviral signal transduction pathway mediated by IFNs. This system is an essential part of the IFN-induced antiviral defense against the encephalomyocarditis virus, reovirus, and the vaccinia virus [44,45]. However, it is not yet known whether the presence of these proteins can prevent the initial transmission of FLUAVs from wild birds to domestic poultry, a key question for poultry health. Uchida et al. [46] suggested the need to elucidate the role of OASL through recombinant HPAI virus infection experiments. A study published in 2024 [47] demonstrated that chicken OASL targets VP2, a key protein of infectious bursal disease virus (IBDV), inducing its degradation through the autophagy pathway and thereby inhibiting IBDV replication. Additionally, a 2022 study [48] revealed that the structure and immune-regulatory functions of chicken OASL are similar to those of mammalian ISG15, suggesting its critical role in the innate immune response of chickens. Nevertheless, no study to date has reported the independent antiviral activity of OASL against HPAI in poultry. The OAS family proteins are known to exhibit antiviral effects against various viruses through two major pathways [44,49–51], but the intracellular antiviral mechanism of OASL requires further study. Our results revealed the OASL gene contains an infection resistance-conferring SNP that enhances the host’s defense against H5N6 infection (Figs. 6, 8B, and 8C). Among the OASL subtype family members, human OASL and mouse OASL2 show the same SNP as the Jeju Island-specific OASL subtype (Fig. 8A). This SNP is positioned close to the OAS-specific motif RPVILDPADP and lies in one of the alpha-helices of human OAS1 [52,53]. Although a direct relationship between this domain and antiviral activity has not been confirmed, further studies on the OASL subtype identified in this research are expected to provide critical insights into the antiviral mechanisms against HPAI.
To exclude interference from other candidate genes, we performed infection experiments using mammalian 3T3 cell lines in which Mx1 and OASL gene variants were individually inserted and expressed. The expression of genes with resistance-associated SNPs enhanced antiviral activity over that of their corresponding reference sequence gene (Fig. 8). These results suggest a need for an integrated analysis of multiple genes that show resistance while accounting for virus type and environmental factors, moving beyond the searches for AI resistance factors that have focused on Mx1 or a single gene.
Unfortunately, we were unable to amplify the virus sufficiently to demonstrate a correlation between H5N1 infection and the antiviral activity of OASL. However, it has been reported that H5N1 infection upregulates the expression of Mx1 and OASL in resistant R1 chickens carrying the resistant-type Mx1 gene (N631) and the BF2/B21 haplotype [54]. This suggests that further experiments with H5N1 may yield results similar to those seen with H5N6 infection in the current study. In addition, given the results of our cell-infection experiments, follow-up research is needed to verify the protective effects of the identified Mx1 and OASL variants in vivo and develop experimental techniques that can easily type each gene using genomic DNA extracted from small amounts of blood or tissue. Our research team has completed the development of a PCR-RFLP method that can efficiently identify OASL SNPs showing antiviral activity against H5N6 and is using it in combination with existing technology that can identify Mx amino acid 631 to genetically characterize chickens. To support the preservation and development of breeds, the experimental results have been shared with the local government agency (Jeju Special Self-Governing Province Livestock Promotion Agency) responsible for maintaining and supplying the ancestral lines of farm chickens used in this study. Collaborative efforts are underway to ensure the thorough management of these breeds and to register their genetic resource information.
In conclusion, we observed that, among the chicken breeds maintained on Jeju Island, individuals with specific SNP combinations in Mx and OASL showed antiviral activity against H5N6. In addition, we suggested the need to characterize candidate innate immunity genes and conduct AI-resistance studies that use combinations of multiple genes and incorporate virus type and environmental characteristics, moving beyond single-gene antiviral activity studies. Based on these results, it is expected that it will be possible to overcome the limitations of preventive vaccines and establish a biological defense system that can block the spread of AI from the wild to humans at the intermediate host while reducing economic losses caused by mass culling of poultry.
