INTRODUCTION
Lactiplantibacillus plantarum (L. plantarum), formally known as Lactobacillus plantarum, stands as one of the most frequently used probiotic strains. Among probiotics, L. plantarum is renowned for possessing one of the largest genomes. Its ability to withstand gastric transit enables easy colonization within the intestines of humans and various other mammals [1]. Thriving in diverse ecosystems, L. plantarum exhibits exceptional probiotic properties and holds promise as a beneficial addition to the livestock industry [2].
In this study, we isolated the L. plantarum strain GA_C_14 from gajami sikhae, a traditional Korean food purchased from a local market in Gangneung, South Korea. Subsequently, whole genome sequencing of L. plantarum strain GA_C_14.was conducted to understand its genomic characteristics, aiming to explore its potential as a probiotic in the livestock industry. L. plantarum strain GA_C_14 was cultured using agar solidified by mixing broth supplemented with L-cysteine and BactoTM Agar (BD Bioscience, Seoul, Korea) in Reinforced clostridial media (BD Bioscience), followed by anaerobic culture at 37°C for 36 hours. The culture was maintained in 25% glycerol solution at -70°C until further use. DNA extraction from the cultured pellet of L. plantarum GA_C_14 was performed using the CTAB method. The Oxford Nanopore Technologies MinION platform at eGnome (Seoul, Korea) was employed to fully sequence the complete genome of the L. plantarum strain GA_C_14. Initially, Native barcoding sequencing (SQK_NBD114.96 V14) was utilized for library preparation, following the manufacturer’s guidelines from Oxford Nanopore Technologies (Oxford, UK). Subsequently, the prepared library was inserted into the MinION MK1b sequencing device (Oxford Nanopore) utilizing a MinION flow cell (MIN114, R10.4.1, Oxford Nanopore), and then verified through the MinKNOW software. A total of 52,921 long read sequences (575,846,933 base pairs) were generated through the Oxford Nanopore sequencing. The Flye assembler v2.9.2 and Canu assembler v1.8 methods were utilized for the de novo assembly. Subsequently, the assembled genome was further refined by employing the Homopolish polisher v0.4.1. The genome assembly’s quality was evaluated employing the Quality Assessment Tool for Genome Assemblies (QUAST) v5.0.2 [3]. Benchmarking Universal Single-Copy Orthologs (BUSCO) v5.4.6 was employed for the quantitative evaluation of genome completeness [4]. The Rapid Annotation using Subsystem Technology (RAST) v2.0 tool was utilized for annotating and predicting protein coding genes, tRNA, and rRNA genes in L. plantarum strain GA_C_14 [5]. The Clusters of Orthologous Groups (COGs)-based EggNOG-mapper v2.0 was utilized for the functional classification of all predicted protein coding genes. Additionally, the BLASTn method with reference to the Virulence Factor Database (VFDB) was used to predict the presence of virulence factors in L. plantarum strain GA_C_14. Identification of antimicrobial resistance genes was performed using the ResFinder v.4.4.0 [6].
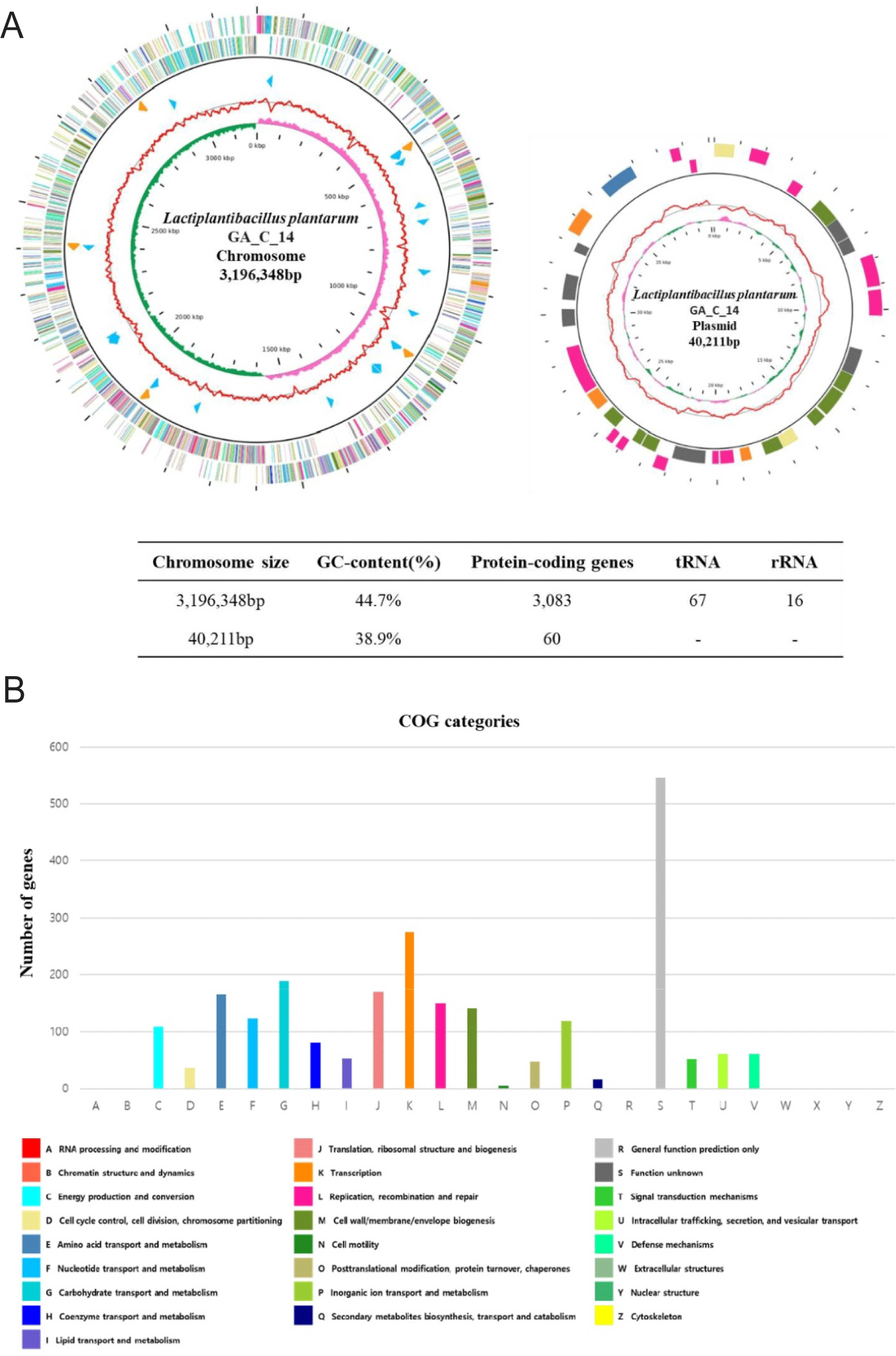
The complete genome of the L. plantarum strain GA_C_14 comprises one circular chromosome spanning 3,196,348 base pairs with a guanine + cytosine (GC) content of 44.7%. Additionally, it contains one circular plasmid measuring 40,211 bp, exhibiting a GC content of 38.9%. Within the genome of L. plantarum strain GA_C_14, a total of 3,083 predicted protein-coding sequences, 67 tRNA, and 16 rRNA genes were identified in. Table 1, Figs. 1A and 1B show the detailed genome features and the map of L. plantarum strain GA_C_14.
L. plantarum strain GA_C_14 possesses genes associated with enzymes crucial for effective vitamin B6 production and usage, as well as for galactooligosaccharides (GOS) synthesis. Specifically, it harbors genes involved in vitamin B6 metabolism: gapB (EC 1.2.1.12), SerC (EC 2.6.1.52), dxs (EC 2.2.1.7), SerA (EC 1.1.1.95), PdxK (EC 2.7.1.35) and PdxH (EC 1.4.3.5), facilitating the production and utilization of vitamin B6 [7]. In swine, Vitamin B6 deficiency can lead to decreased appetite and hindered growth [8]. Additionally, the L. plantarum strain GA_C_14 carries the beta-galactosidase (EC 3.2.1.23) enzyme, crucial for synthesizing GOS [9]. This enzyme catalyzes the transgalactosylation reaction in GOS synthesis. Studies have demonstrated that supplementing swine diets with GOS can promote swine growth and enhance intestinal immune status [10]. Therefore, this characteristic suggests that the L. plantarum strain GA_C_14 could be used as a potential probiotic candidate for application in the swine industry. Notably, the complete genome of L. plantarum strain GA_C_14 does not contain antibiotic resistance genes or virulence factors. In conclusion, our study underscores the potential of L. plantarum strain GA_C_14 as a functional probiotic candidate, showing its capacity to positively impact growth outcomes in the swine industry.
