INTRODUCTION
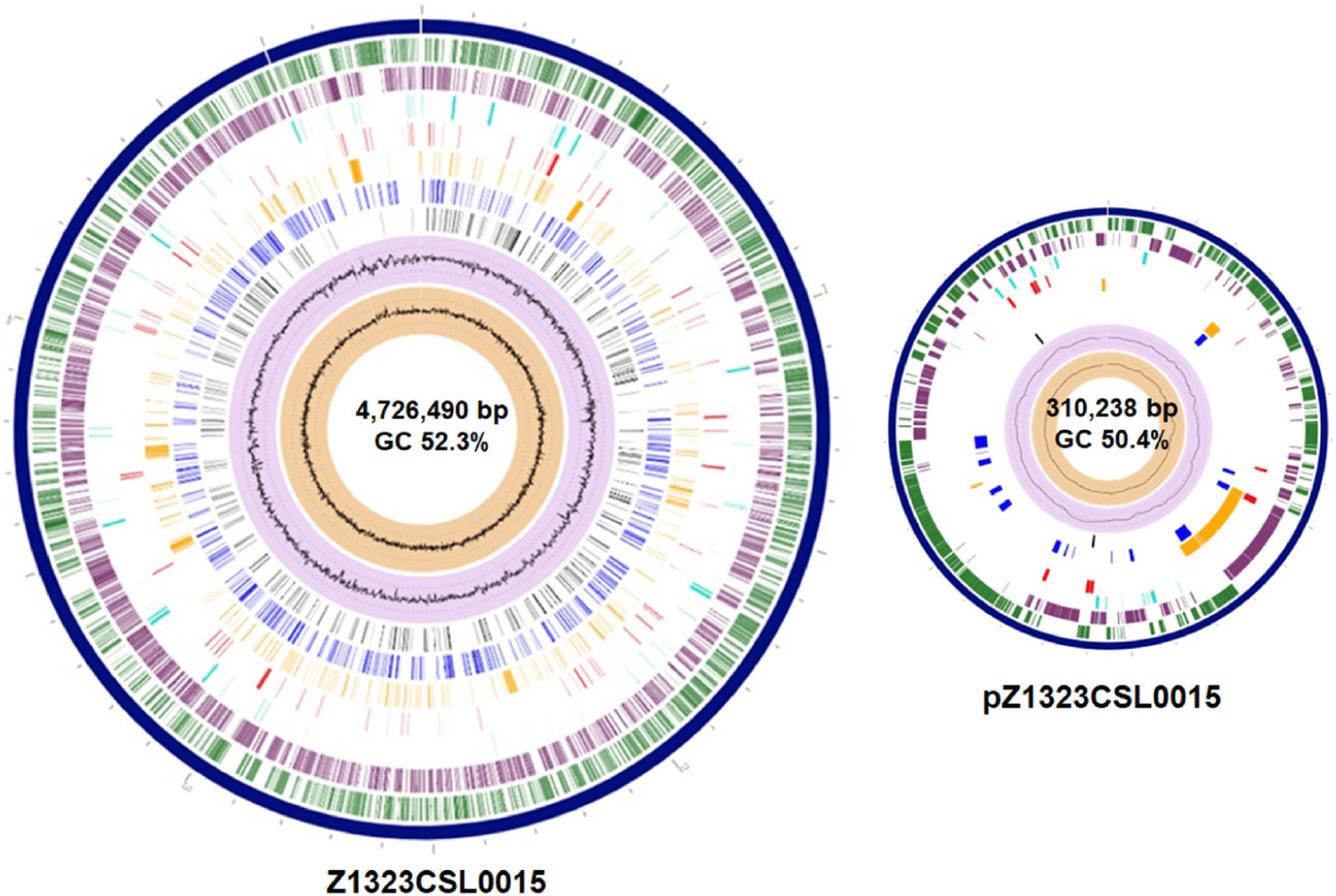
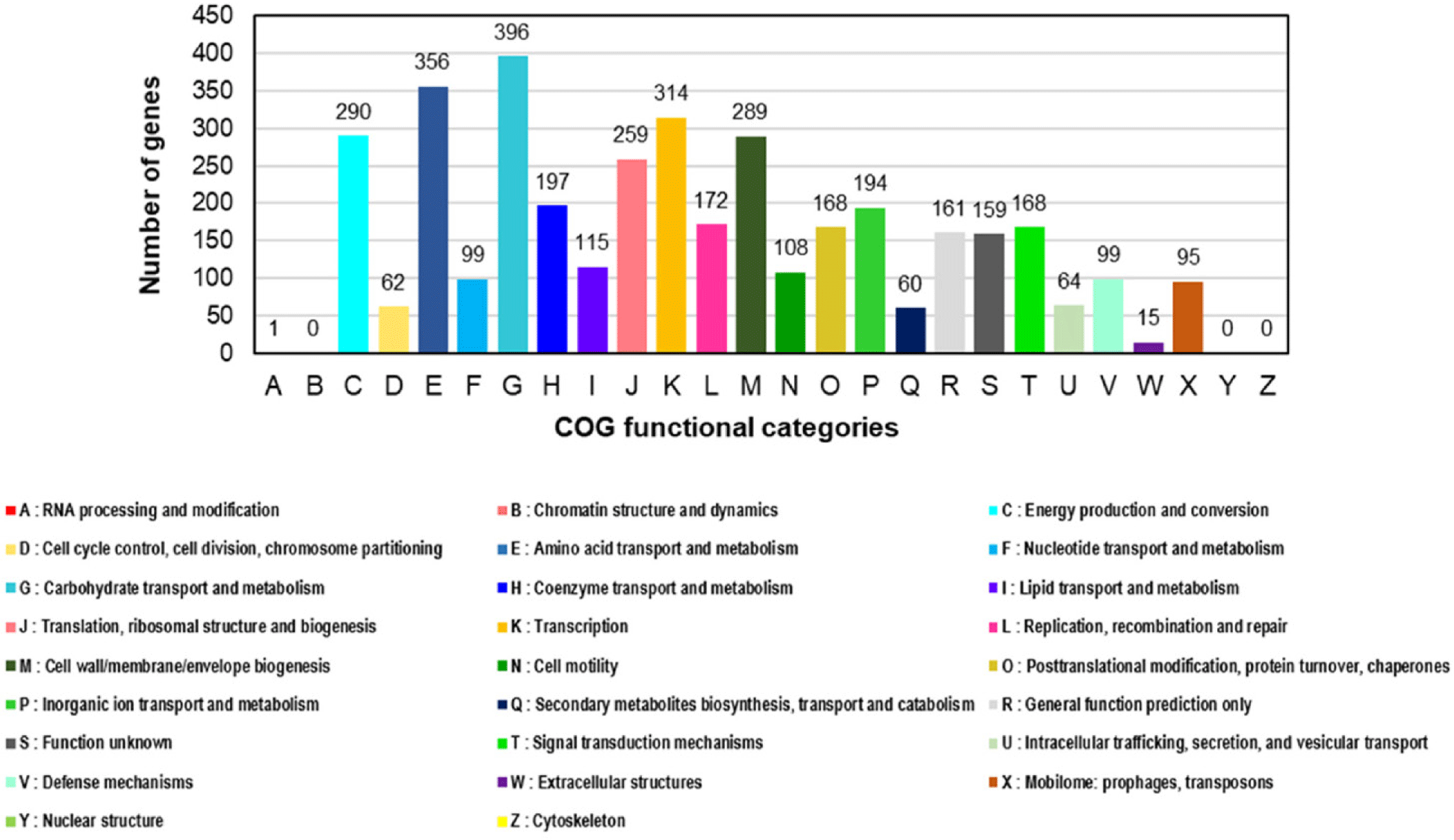
Salmonella spp. are globally recognized as acute pathogens that cause gastroenteritis in humans [1–4]. Some Salmonella serovars have been detected in the food production stage and may colonize poultry and swine farms [2,3]. Salmonella Infantis has emerged as the fourth most prevalent serovar associated with human illness after S. Enteritidis, S. Typhimurium, and S. Typhimurium monophasic variants [4]. Salmonella Infantis strain Z1323CSL0015 was isolated from a cloacal swab specimen of a 21-day-old broiler chicken from Korea. This strain was cultivated according to the Food Code (Ministry of Food and Drug Safety). First, the swab sample was suspended in Rappaport-Vassiliadis Salmonella (RVS) enrichment broth (Difco, Tucker) and cultivated under aerobic conditions at 42°C for 48 h. The culture was inoculated on Rambach agar (CHROMagar) and incubated at 37°C for 24 h. Resulting well-isolated violet colonies were consecutively cultivated on MacConkey agar (Difco). Genomic DNA of the Z1323CSL0015 strain was extracted using the DNeasy Blood and Tissue kit (QIAGEN) according to the manufacturer’s instructions. The genome sequencing of S. Infantis strain Z1323CSL0015 was conducted in our laboratory and Macrogen for long- and short-read sequencing. A MinION sequencer (Oxford Nanopore Technologies) equipped with a Flow Cell R10 Version (Oxford Nanopore Technologies) was used to generate long-read sequences, whereas short-read sequences were obtained using HiSeq X (Illumina). The Illumina and Nanopore sequencing platforms produced 12,500,648 and 1,248,961 reads corresponding to the sequencing depth of 287.6× and 330.0×, respectively. Short-read raw sequences were processed for trimming adapters/primers and low-quality sequences using Trimmomatic (v. 0.39) [5]. The long-read raw products were treated to trim low-quality sequences with Filtlong (v. 0.2.0) and adapter sequences with Porechop (v. 0.2.4) [5]. Hybrid de novo assembly was performed using Unicycler (v. 0.4.9b), followed by polishing with Pilon (v. 1.21) [5]. All the predicted protein coding genes were assigned to the Clusters of Orthologous Groups (COGs) database [6] using COGclassifier version 1.0.5 (https://github.com/moshi4/COGclassifier). Potential virulence factors and antimicrobial resistance genes in the Z1323CSL0015 strain were predicted using the analysis algorithms in the bacterial and viral bioinformatics resource center (BV-BRC) [7]. The complete genome of the Z1323CSL0015 strain consisted of one circular chromosome (4,726,490 bp, 52.3% guanine-cytosine [GC] content) and one plasmid designated as pZ1323CSL0015 (310,238 bp, 50.4% GC content) (Table 1 and Fig. 1). The best match among the similar strains based on 16S rRNA gene sequences was Salmonella enterica subsp. enterica Serovar Infantis strain CVM N17S1509 (GenBank Nos. CP052817 and CP052818) which also contained a mega-plasmid, pN17S1509 [8]. Average nucleotide identity (ANI) value was 99.97% between the genomes. The complete genome of strain Z1323CSL0015 comprised 5,087 protein coding sequences (CDSs) and 105 non-coding genes (22 rRNA and 83 tRNA genes). A total of 3,841 proteins were classified into functional categories based on the COGs database. The most abundant COG categories were carbohydrate transport and metabolism (category G, 396 genes, 10.3%), followed by amino acid transport and metabolism (category E, 356 genes, 9.3%), transcription (category K, 314 genes, 8.2%), energy production and conversion (category C, 290 genes, 7.6%), and cell wall/membrane/envelope biogenesis (category M, 289 genes, 7.5%) (Fig. 2). The plasmid of Z1323CSL0015 strain contained various antimicrobial resistance genes such as blaCTX-M-65 (cefotaxime), aph(4)-la (atypical aminoglycoside), aac(3)-V (gentamicin and netilmicin), aadA (streptomycin and spectinomycin), tetC (tetracycline), dfrA14 (trimethoprim), sul1 (sulfonamide), and qacEdelta1 (quaternary ammonium compounds). These genes were also found in pN17S1509 mentioned above except for blaCTX-M-65. Salmonella Infantis carrying multidrug-resistant pESI (plasmid for emerging S. Infantis)-like mega plasmids have been reported in many countries [2]. In a view of horizontal gene transfer, the plasmid of Z1323CSL0015 could be disseminated among the serotypes conferring antibiotic resistance. Therefore, our genomic information might be useful for the surveillance of plasmid-mediated antibiotic resistome.