INTRODUCTION
Pigs are one of the major livestock, with pork accounting for more than a quarter of the total protein consumed worldwide and approximately 35% of the total meat production [1]. Pigs are recognized as important livestock, and various studies are being conducted to increase their productivity. The health status of pigs is influenced by a combination of multiple factors, including genetics, environmental stress, pathogen infection, and nutrition [2–4]. Among these factors, nutrition is particularly related to gut health, and numerous studies have focused on enhancing gut health in pigs. For example, positive indicators related to gut health, such as reduced diarrhea incidence, increased tight junction protein gene expression, and improved intestinal morphology, were identified when plant-derived oils rich in n-3 polyunsaturated fatty acids were fed to weaned piglets as feed additive [5]. In contrast, an in vitro study using the pig intestinal epithelial cell line, IPEC-J2, examined the effects of functional nutrients on gut health. For example, acetate and propionate enhance cell viability and gut barrier integrity [6].
Recently, because of animal welfare issues, methods that can replace animal experiments have attracted considerable attention. However, in vitro studies in domestic animals are limited. Organoid culture systems can mimic and reproduce tissue functions and properties. For example, organoid systems are highly similar to living organisms and can be applied in genetic engineering, making them economically and efficiently suitable for high-throughput screening [7]. In addition, organoids have the characteristics of cell populations related to organs, which enables the study of interactions with factors related to these organs. A representative example is the intestinal organoid-based co-culture system used in mechanistic studies [8]. In a study by Hou et al., an organoid-based co-culture model with lamina propria immune cells isolated from the intestine was developed, and the immune cell-epithelium regulatory mechanism of Lactobacillus reuteri, a well-known probiotic, was investigated. Thus, organoid-based co-culture models are recognized as advanced tools with the potential for in vitro research on biological processes [9,10]. Intestinal organoids can be generated using embryonic, pluripotent, and adult stem cells [11]. These stem cells can be cultured under appropriate culture medium conditions without a specific feeder cell [12]. For example, isolated crypts, which contain adult stem cells, undergo self-renewal, organization, morphogenesis, and differentiation within the crypt-villus structure [13]. Crypt-derived intestinal organoids exhibit structural and functional similarities to the gut.
Recently, various intestinal organoids have been developed for livestock, including cattle, sheep, chicken, and pigs [14–17]. Many studies have investigated intestinal diseases induced by pathogenic microbes and viruses in pig gut organoids. Disease-inducing microbes such as Salmonella typhimurium and Toxoplasma gondii can directly infect pig organoids [18]. In a study by Li et al., a transmissible gastroenteritis virus, a pig enteric coronavirus, was found to infect pig jejunal organoids. In a previous study, apical-out and two-dimension (2D) culture methods were used due to structural limitations in three-dimension (3D) culture, which is the basic organoid culture method [19]. Pathogens mainly infect the intestinal lumen, and most nutrient uptake and sensing occurs in the intestinal lumen, resulting in metabolic processes. Thus, the 2D organoid model has several advantages as an in vitro research platform because it can simulate phenomena occurring in the intestinal lumen.
Although studies on pig intestinal organoids have been actively conducted, the characteristics of these platforms remain unclear. Therefore, this study aimed to compare the characteristics of the existing pig intestine in vitro platform with pig organoids, and to establish a 2D organoid model for better intestinal epithelial cell research. A 2D organoid model was developed to simulate external exposure of the lumen using pig 3D organoids, and the physiological and nutritional characteristics were compared. Our results suggest the possibility of using the 2D pig organoid model in nutritional research.
MATERIALS AND METHODS
IPEC-J2, a pig intestinal cell line, was kindly provided by Prof. Yun of Seoul National University. IPEC-J2 cells were cultured in a 90 mm cell culture dish (SPL Life Sciences) using DMEM/F12 medium (Thermo Fisher Scientific), supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific) at 37°C in a humidified 5% CO2 atmosphere. For RNA extraction and immunofluorescence staining, 5 × 105 cells were seeded to 6-well plates (SPL Life Sciences) and 35 mm confocal dishes (SPL Life Sciences) for 2 days.
R-spondin 1 and Wnt-3a are critical factors in pig intestinal organoids. To procure them, two cell lines expressing the target proteins were used in cell culture-conditioned media. Conditioned media were prepared as described previously [20]. Briefly, R-spondin 1-expressing HEK293T cells and L Wnt-3A cells (CRL-2647™, ATCC) were cultured in a 90 mm cell culture dish using DMEM medium (Thermo Fisher Scientific), supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, with selective antibiotics for 2–3 passages. The selective antibiotics used were: 0.2 mg/mL Hygromycin B (Thermo Fisher Scientific) for HEK 293T cells and 0.4 mg/mL G-418 (Thermo Fisher Scientific) for L Wnt-3A cells. To obtain a high concentration of conditioned media, the cells were initially cultured (5 × 105 cells in a 90 mm cell culture dish) for 7 days using advanced DMEM/F12 medium (Thermo Fisher Scientific) with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Briefly, cells were grown for 4 days (approximately 80%–90% confluence at this point) in harvest media (first batch). The medium was replaced with fresh culture medium, and the cells were cultured for another 3 days in the harvest media (second batch). Finally, the first and second batches were mixed and filtered in a 1:1 ratio and stored at –20°C until further use.
In this study, three jejunal fragments were harvested from 3-week-old weaned piglets, all of which were healthy and asymptomatic. For crypt isolation, 3–4 cm of the gut tissue was harvested and opened longitudinally. To remove luminal contents and mucus, they were gently scraped using slide glass and vigorously washed with phosphate-buffered saline (PBS; Thermo Fisher Scientific). The gut tissues were then cut to 0.5 × 0.5 cm2 pieces and transferred to a crypt isolation solution containing 30 mM ethylene-diamine-tetra acetic acid (EDTA; Thermo Fisher Scientific) and 1 mM DL-dithiothreitol (DTT; Sigma-Aldrich). The tissue fragments were incubated for 30 min on ice in a horizontal shaking incubator at 100 rpm. After incubation, the tissue fragments were transferred to a cold crypt washing buffer containing 54.9 mM D-sorbitol (Sigma-Aldrich) and 43.4 mM sucrose (Sigma-Aldrich) in PBS, and gently shaken for 2 min to release the crypts. To obtain pure crypts, the supernatant was transferred to a new tube using 100 μm cell strainer (SPL Life Sciences) and centrifuged at 200×g for 5 min. The crypt pellet was resuspended in advanced DMEM/F12 medium and counted for pig intestinal organoid culture.
The counted pig jejunal crypts were mixed with advanced DMEM/F12 and Matrigel (Corning) in a 1:1 ratio. The mixture was seeded into a 96-well cell culture plate (SPL Life Sciences) at a concentration of 5 crypts/μL (total volume: 4 μL). The plate was then incubated for 30 min in a cell culture incubator to solidify the Matrigel mixture. Next, 100 μL of pig intestinal organoid culture medium was added to each well, and medium composition was as follows: advanced DMEM/F12 supplemented with 1x N2 supplement (Thermo Fisher Scientific), 1x B27 supplement (Thermo Fisher Scientific), 2 mM GlutaMAX™ Supplement (Thermo Fisher Scientific), 10 mM nicotinamide (Sigma-Aldrich), 1 mM N-acetylcysteine (Sigma-Aldrich), 10 mM HEPES (Thermo Fisher Scientific), 100 μg/mL Primocin™ (InvivoGen), 50 ng/mL recombinant murine EGF (PMG8041, Thermo Fisher Scientific), 100 ng/mL recombinant murine Noggin (250-38, PeproTech), 10% R-spondin 1 conditioned media, 50% Wnt-3a conditioned media, 10 μM SB 202190 (Sigma-Aldrich), 0.5 μM A 83-01 (Sigma-Aldrich), 2.5 μM CHIR99021 (Sigma-Aldrich), and 10 μM Y-27632 (Selleckchem). Pig organoids were cultured for 4 days, and the medium was replaced on day 2. To prevent anoikis in pig organoids, Y-27632 was added for the first two days only.
To maintain pig organoids and develop 2D monolayer organoid, 4-day-cultured organoids were sub-cultured. Matrigel was dissociated using cell recovery solution (Corning) at 4°C for 30 min using an orbital shaker with slow shaking (60 rpm). The organoid-containing supernatant was collected and centrifuged at 200×g for 5 min. Pig organoid pellets were resuspended in advanced DMEM/F12 and physically pipetted. Organoid fragments were counted and cultured on a new plate using the methods described above.
Sub-cultured organoid fragments were seeded into a 96-well cell culture plate at 12.5 crypts/μL concentration (total volume: 4 μL) and cultured for 2 days. Short-cultured pig organoids were passaged for the sub-culture method, and organoid pellets were incubated with TrypLE Express Enzyme solution (Thermo Fisher Scientific) with occasional pipetting for 10 min in a cell culture incubator. Pig organoids dissociated into single cells were centrifuged at 800×g for 5 min and counted. The single cells were resuspended in 2D pig intestinal organoid culture medium with 20% (v/v) fetal bovine serum added to the pig intestinal organoid culture medium, and seeded at 150,000 cells/cm2 in pre-coated plates. The pre-coating process was carried out by incubating 2% (v/v) Matrigel with advanced DMEM/F12 medium (50 μL for 96-well cell culture plate) for 1 h in a cell culture incubator. Before single cell seeding, the coating solution was removed and the cells were washed once with advanced DMEM/F12.
Total RNA of all samples, including IPEC-J2 cells, 3D organoids, and 2D organoids, was extracted using TRIzol™ Reagent (Thermo Fisher Scientific). The 0.5 μg of RNA was used for cDNA synthesis using the AccuPower® RT PreMix (Bioneer). qRT-PCR was performed using a QuantStudio 1 Real-Time PCR system (Applied Biosystems) and the following conditions were used: 50°C for 2 min, 95°C for 15 min, 95°C for 20 s, and 60°C for 40 s (40 cycles), followed by melting curve analysis. GAPDH was used for normalization of relative gene expression, and the expression level was calculated using the 2-ΔCT method [21]. The primer sequences for the target genes used in this study are presented in Table 1.
Inverted and confocal microscopes were used to obtain organoid images. A Nikon Eclipse Ts2R microscope (Nikon) was used to obtain day-to-day 3D and 2D organoid images. Immunofluorescent staining was performed to compare the expression of intestinal epithelial cell markers in IPEC-J2 cells and 3D organoids, and images were obtained using a confocal microscope. Briefly, the cells were fixed in 2% paraformaldehyde (Biosesang) for 30 min at room temperature. After washing with PBS, cells were permeabilized with 1% Triton X-100 (Biosesang) for 30 min at room temperature. The samples were washed with PBS and blocked with a blocking buffer (10% goat serum and 0.5% Triton X-100) overnight at room temperature. The primary antibodies rabbit anti-Muc2 (27675-1-AP, ProteinTech) and mouse anti-ChgA (sc-393941, Santa Cruz Biotechnology) were diluted at 1:50 and 1:100, respectively, with the blocking buffer. The samples were then incubated for 4 h at room temperature and washed 5 times using the blocking buffer. After washing step, the secondary antibodies, goat anti-rabbit IgG Alexa Fluor 488 (A-11034, Thermo Fisher Scientific) for Muc2 and goat anti-mouse Alexa Fluor 488 for ChgA (A-11029, Thermo Fisher Scientific), were diluted at 1:500 using the blocking buffer. The samples were incubated for 1 h at room temperature in the dark and washed 10 times using the blocking buffer. The samples were stained with Alexa Fluor 555 phalloidin (Thermo Fisher Scientific; 1:200 diluted in PBS) and Hoechst 33342 (Thermo Fisher Scientific; 1:500 diluted in PBS). Both staining processes were performed sequentially, and the samples were incubated in the dark for 30 min at room temperature and washed with PBS. After staining, IPEC-J2 cells and 3D organoids were subjected to confocal microscopy (K1-Fluo; Nanoscope Systems).
To assay fatty acid uptake, C1-BODIPY-C12 (Thermo Fisher Scientific) was used on 3D and 2D organoids, with minor modifications [20]. For the 3D organoid assay, Matrigel was solubilized in a cell recovery solution using the above method. They were resuspended in a solution of 1 μM C1-BODIPY-C12 with 10% fatty acid-free bovine serum albumin solution and incubated in an ultra-low attachment 24-well cell culture plate (Corning) for 30 min in a cell culture incubator. For the 2D organoid assay, the medium was removed, and they were incubated for 30 min in a cell culture incubator with the same BODIPY solution. The 3D and 2D organoids were fixed in 2% paraformaldehyde and stained with Hoechst 33342 for image analysis. The intracellular fluorescent signal (fluorescent size and intensity) was quantified using NIS-Elements Basic Research software (Nikon).
The lipid mixture (LM; Sigma-Aldrich) was used to treat both 3D and 2D organoids. It contains non-animal derived fatty acids (2 μg/mL arachidonic acid and 10 μg/mL each linoleic, linolenic, myristic, oleic, palmitic, and stearic acids), 0.22 mg/mL cholesterol from New Zealand sheep′s wool, 2.2 mg/mL Tween-80, 70 μg/mL tocopherol acetate, and 100 mg/mL Pluronic F-68 solubilized in cell culture water. Before harvesting the 3D and 2D organoid samples (on day 4 for 3D organoids and on day 2 for 2D organoids), they were treated with 2% LM (v/v) (in each organoid culture medium) for 12-hours. All LM-related organoid experiments, including qRT-PCR, MitoTracker staining, and ADP:ATP ratio assays, were performed using the same method.
To stain the mitochondria of organoids, MitoTracker™ Green FM (Thermo Fisher Scientific) was used. MitoTracker was prepared as a 1 mM stock in dimethyl sulfoxide according to the manufacturer’s instructions. The 3D and 2D organoids were washed once with advanced DMEM/F12, and MitoTracker (final concentration of 100 nM) and Hoechst 33342 (final 1:500 dilution) were added to each organoid culture medium. They were then incubated for 30 min in a cell culture incubator and washed once with advanced DMEM/F12. The stained images were immediately obtained using a confocal microscope.
ADP:ATP ratios of 3D and 2D organoids were analyzed according to the bioluminescence method using a SpectraMax iD5 microplate reader (Molecular Devices). The assay was conducted using a commercial ADP:ATP Ratio Assay Kit (Abcam) according to the manufacturer’s instructions. Briefly, Matrigel was removed from 3D organoids using a cell recovery solution, and single cells were obtained using the TrypLE Express Enzyme solution. After centrifugation, incubation was performed at room temperature for 5 min using the Nucleotide Releasing Buffer of the kit (200 μL per well in a 96-well plate). 2D organoids were washed once with plain advanced DMEM/F12 and incubated at room temperature for 5 min in an equal volume of Nucleotide Releasing Buffer. The remainder of the assay was performed according to the method recommended in the kit using a white 96-well plate (SPL Life Sciences).
The experimental data from three to five independent experiments were pooled and presented as mean ± SD. For fatty acid absorption assay, five representative organoid images (3D organoids) and five position images (2D organoids) were randomly selected within each three independent experiments, and fluorescence area and intensity were measured within selected images (average 4.6 and 9.6 fluorescence signal areas for 3D and 2D organoids, respectively). To determine whether the data were normally distributed, the Shapiro-Wilk test was performed using Prism 8 software (GraphPad). Abnormally distributed data were further analyzed using a two-tailed Mann-Whitney test, and normally distributed data were further analyzed using a two-tailed unpaired t-test. The significant difference between groups was considered at p < 0.05, and the tendency was considered at 0.05 < p < 0.10.
RESULTS
First, the characteristics of the IPEC-J2 cell line, a well-known in vitro pig epithelial cell platform, and pig small intestinal 3D organoids were compared. To determine the presence of several types of epithelial cells, mRNA expression levels of epithelial cell marker genes (LGR5 for crypt-base columnar cells, LYZ for Panth cells, MUC2 for goblet cells, CHGA for enteroendocrine cells, and ALPI for enterocytes) were compared. In 3D pig organoids, LYZ and ALPI showed significantly higher gene expression than IPEJ-J2 cells. In addition, MUC2 and CHGA were expressed in 3D pig organoids, but not in the IPEC-J2 cell line (Fig. 1A). LGR5 was similarly expressed in the organoids and IPEC-J2 cells. Immunofluorescence staining confirmed the presence of MUC2 and CHGA at the protein level. MUC2 and CHGA expressions were observed in 3D pig organoids, but not in IPEC-J2 cells (Fig. 1B and 1C). These data suggest that pig epithelial cell lines have limitations as in vitro research platforms for studying pig epithelial cells.
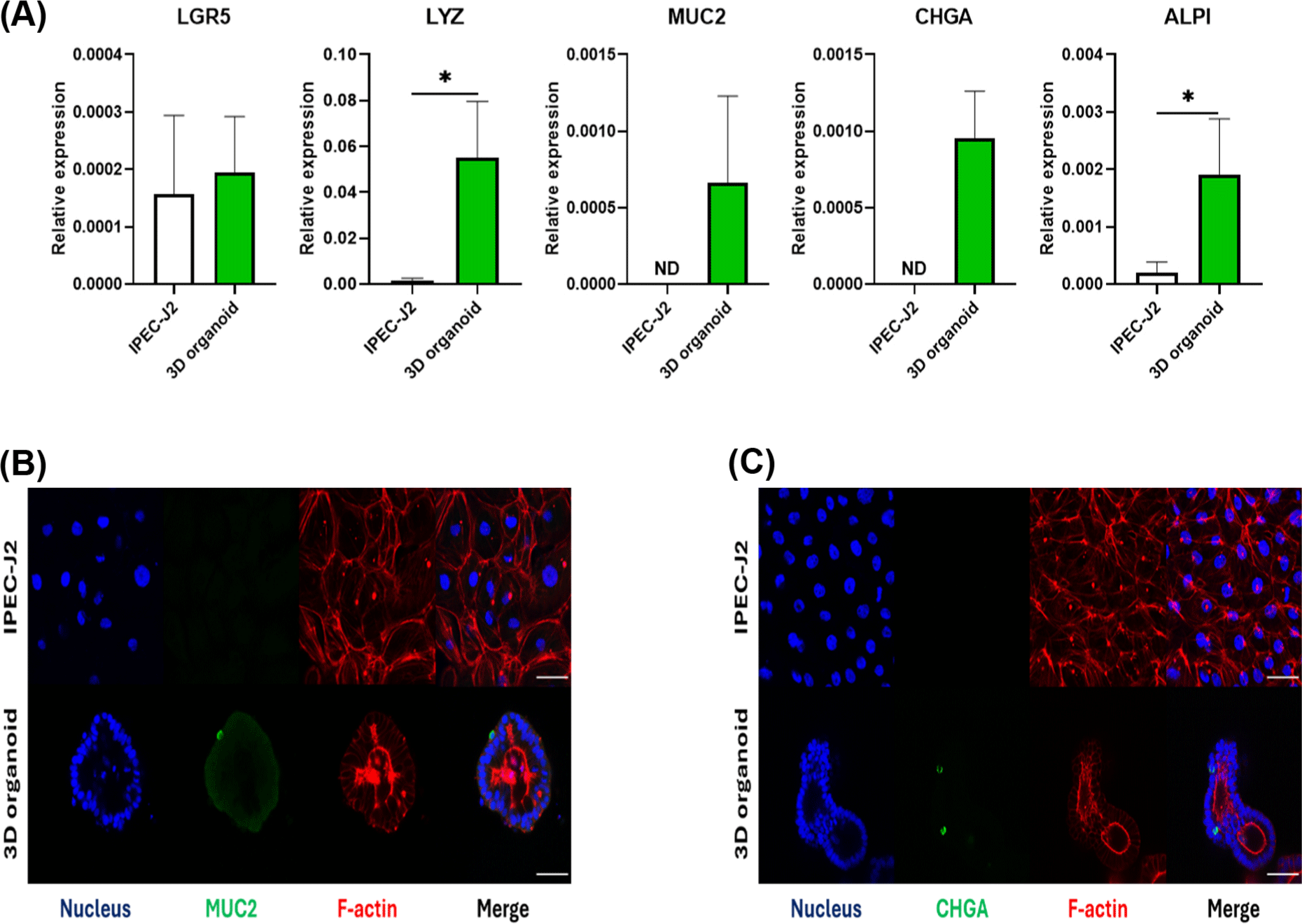
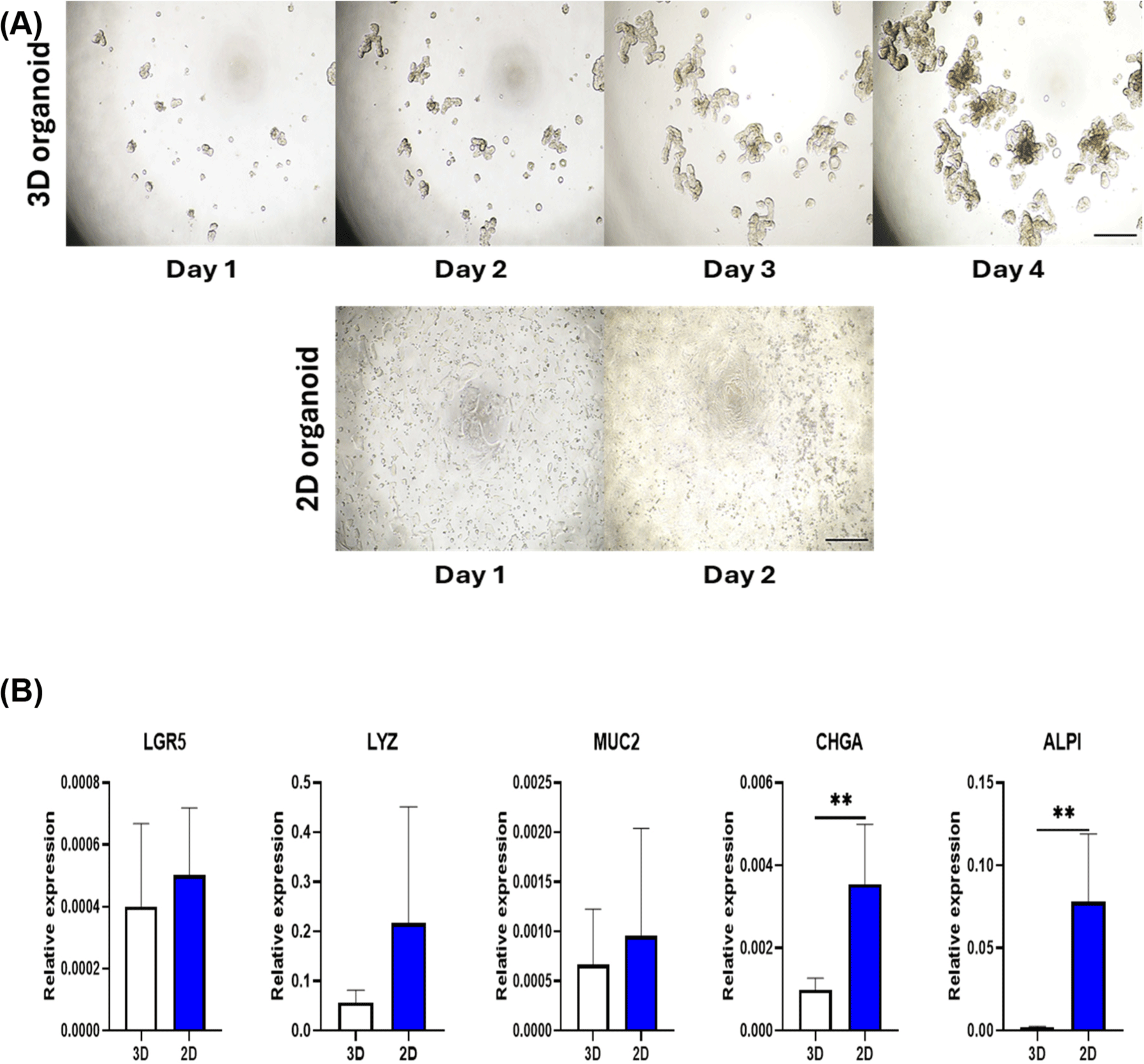
Organoids are normally cultured 3D condition, making it difficult to reproduce phenomenon occurring in the intestinal lumen. As most nutrition-related phenomena occur in the intestinal lumen, a 2D monolayer pig organoid model that can expose the apical membrane was developed in this study. The 3D organoids were fully grown by day 4 by culturing approximately 20 sub-cultured organoid fragments. 2D organoids were seeded with approximately 50,000 single cells from sub-cultured 3D organoids and showed more than 90% confluence on day 2 (Fig. 2A). To compare the expression levels of intestinal epithelial cell marker genes in two fully developed organoids, qRT-PCR was conducted using epithelial cell marker genes. There were no significant differences in LGR5, LYZ, and MUC2 between 3D and 2D organoids. However, 2D organoids had significantly higher expression of CHGA and ALPI than 3D organoids (Fig. 2B). Collectively, these results suggest that, in addition to the structural properties of the 2D organoid model, intestinal epithelial cell marker gene expression differs from that of 3D organoids.
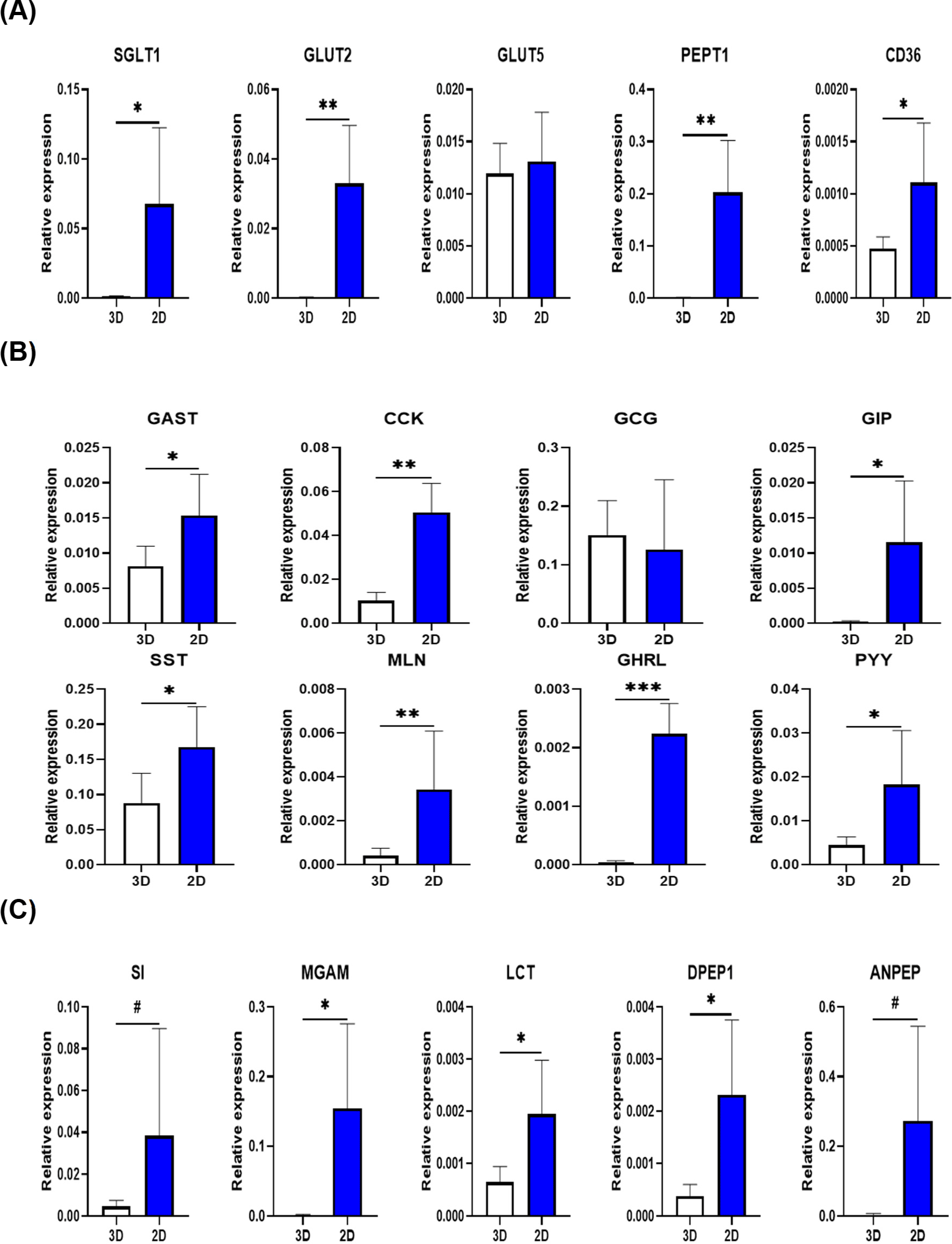
Because high expression of CHGA and ALPI, which are important for nutritional physiological responses, was observed in the 2D organoid platform, nutritional function-related gene expression was further compared. To characterize the nutrition-related functions of pig 2D intestinal organoids, the gene expression levels of small intestinal nutrient transporters, gastrointestinal hormones, and brush border enzymes were compared with those in 3D organoids. Except for GLUT5, a fructose transporter, most nutrient transporters (SGLT1: sodium/glucose transporter, GLUT2: glucose transporter, PEPT1: peptide transporter, and CD36: fatty acid transporter) showed higher gene expression levels in 2D organoids than in 3D organoids (Fig. 3A). Gastrointestinal hormones are mainly secreted by enteroendocrine cells and these hormones are classified into families based on structural homology [22,23]. In this study, the gastrin family (GAST and CCK), secretin family (GCG and GIP), somatostatin family (SST), motilin-ghrelin family (MLN and GHRL), and PP-fold family (PYY) were investigated. Among the various hormone-encoding genes, GCG was not significantly different between 3D and 2D organoids. However, the expression of other hormone-related genes examined in this study was significantly higher in 2D organoids than in 3D organoids (Fig. 3B). The gene expression of brush border enzymes secreted by enterocytes was compared between the two types of pig organoids. The carbohydrate-related (SI, MGAM, and LCT) and peptide-related (DPEP1 and ANPEP) enzyme genes were significantly highly expressed or tended on 2D organoids than in 3D organoids (Fig. 3C). Collectively, these results suggest that 2D organoids have higher functional gene expression for nutritional physiological responses.
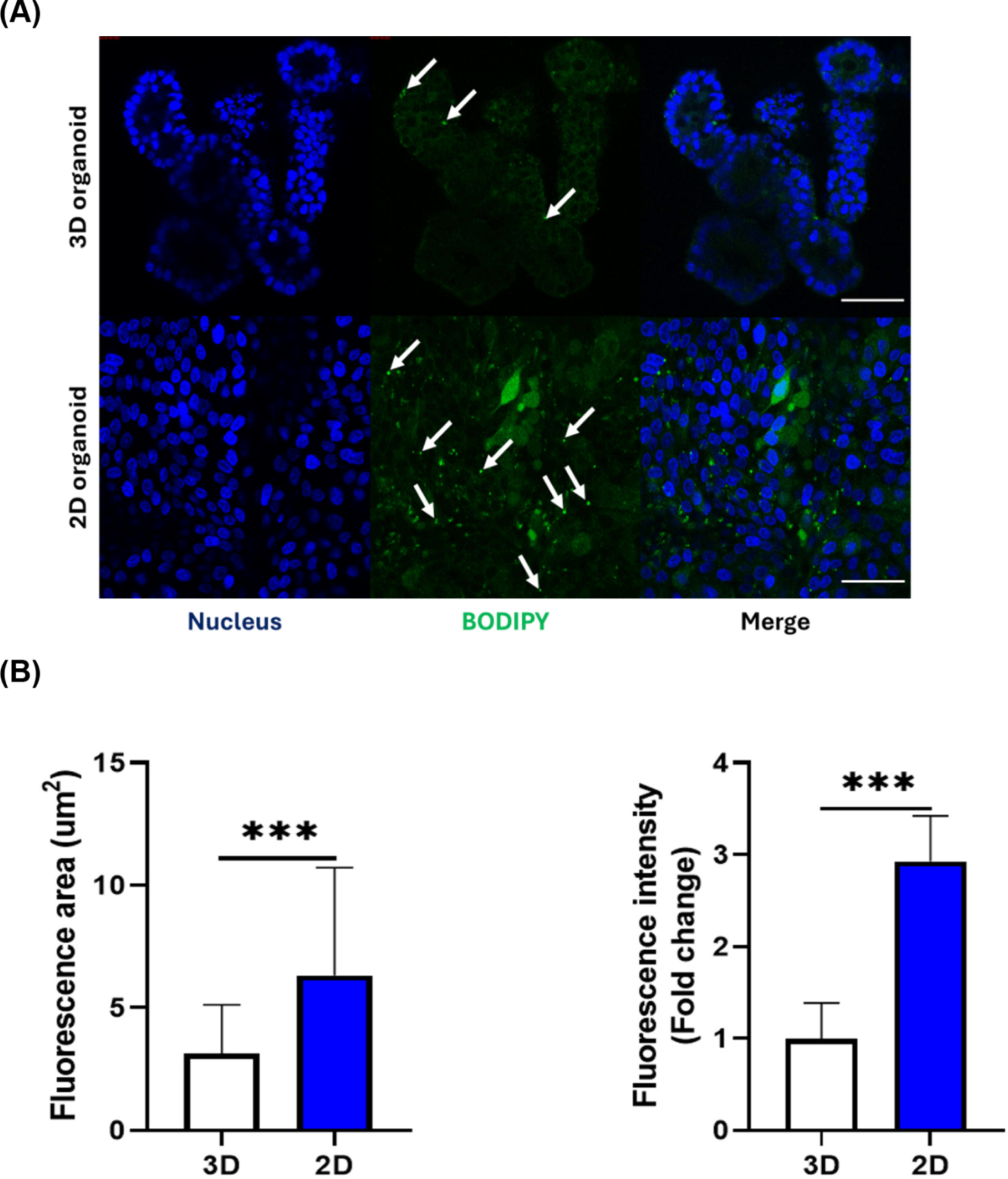
To assess nutrient uptake in 3D and 2D pig intestinal organoids, we used fatty acids as one of the nutrients absorbed by the pig small intestine. To compare the efficiency of fatty acid uptake in each pig organoid model, 3D and 2D organoids were treated with fluorescent fatty acids (Fig. 4A). On average, more lipid droplets were present in 2D organoid images than in 3D organoids. Consistent with this, a larger fluorescence area and brighter fluorescence intensity were observed in 2D organoids than in 3D organoids (Fig. 4B). These data suggest that 2D pig organoids uptake nutrients more efficiently than 3D organoids.
Nutrients are absorbed by intestinal epithelial cells, and nutritional physiological phenomena occur through specific receptors and binding proteins. To evaluate the influence of nutrients, especially fatty acids, LM containing various long-chain fatty acids (LCFAs) was treated to 3D and 2D organoids, and selected LCFA-responsive gene expression was investigated. There was no difference in the gene expression of fatty acid-related receptors (FFAR1 and FFAR4) and binding proteins (FABP1, FABP2, and FABP5), which are associated with enteroendocrine cells and enterocytes, in 3D organoids. However, in 2D organoids, a significant increase and increase tendency in the gene expression of FFAR1 and FFAR4 was observed after LM treatment (Fig. 5A and 5B).
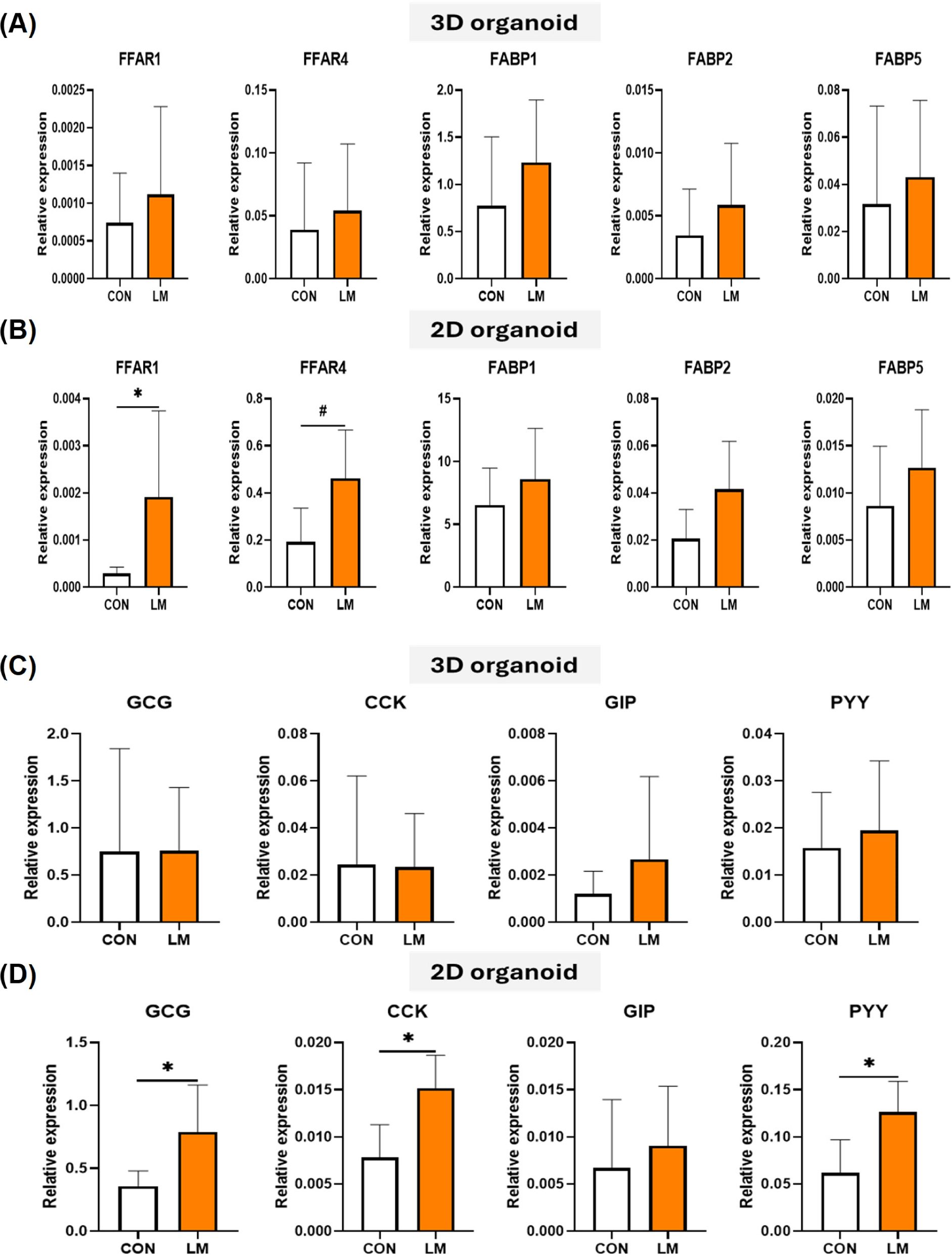
Nutrients present in the intestinal lumen or absorbed by enterocytes may act on enteroendocrine cells to regulate gastrointestinal hormone secretion. Some hormones secreted by the small intestine respond to fatty acids. In this study, major fatty acid-responsive hormone genes, such as GCG, CCK, GIP, and PYY were examined after LM exposure (Fig. 5C and 5D). Treatment of 3D organoids with LM showed no difference in hormone genes, but significant increases in the expression of hormone genes other than GIP were observed in 2D organoids. Overall, these results indicate that the pig 2D organoid model can mimic the nutrient-induced responses occurring in small intestinal epithelial cells, and that 2D organoids are more responsive to lipid molecules than 3D organoids.
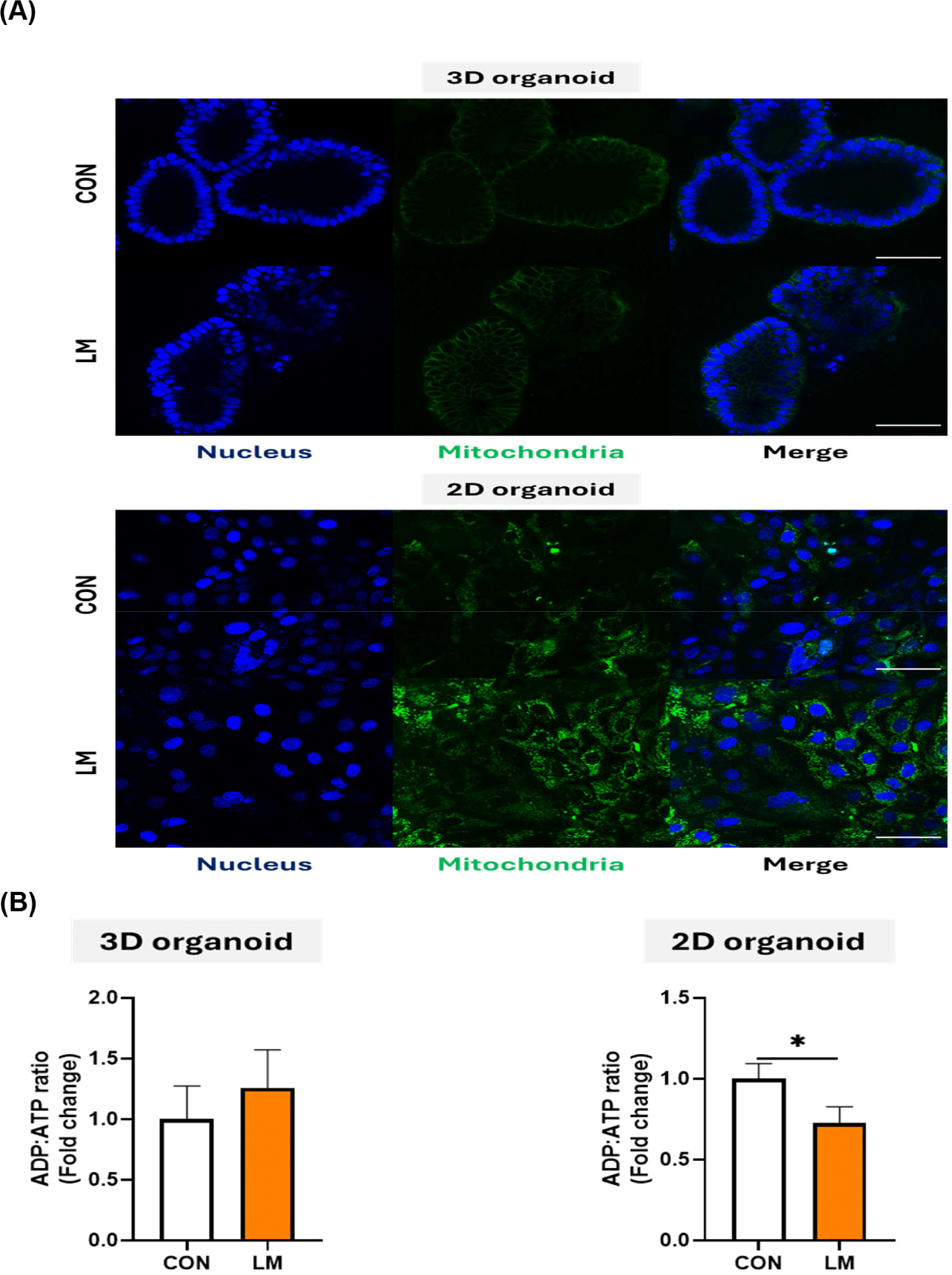
Fatty acids absorbed by intestinal epithelial cells are oxidized by mitochondrial activity to produce energy via ATP synthesis. To assess fatty acid-induced mitochondrial activity and ATP patterns in organoids, we confirmed the mitochondria mass and ADP:ATP ratio in LM-treated 3D and 2D organoids. When the two types of organoids were compared using MitoTracker staining, there was no difference in fluorescence intensity between the control and LM groups of 3D organoids. However, in 2D organoids, strong fluorescence intensity was observed in the LM group compared with that in the control group (Fig. 6A). Next, the intracellular ADP:ATP ratios of 3D and 2D pig intestinal organoids were measured. There was no significant difference between the control and LM groups in 3D organoids. However, in 2D organoids, the ADP:ATP ratio was significantly reduced by LM treatment (Fig. 6B). These results indicate that the 2D organoid model has a more active metabolic response to fatty acids, such as the conversion of ADP to ATP, than the 3D organoid model.
DISCUSSION
The gut is a specialized tissue with multiple functions that interacts with the external environment. The epithelial cell layer of the gut plays an important role in the first physical barrier and immune function against harmful external factors, such as pathogens, viruses, and toxins [24]. The small intestine, a part of the gut, consists of various epithelial cell types. The major cell types include crypt-resident stem cells, Paneth cells, goblet cells, enteroendocrine cells, and enterocytes [25]. Intestinal epithelial cells are continuously regenerated at short intervals, and this phenomenon is due to stem cells differentiating into various cell types through progenitor cells [26]. For regulation of intestinal epithelial cells, the function of intestinal stem cells plays a key role, and the LGR5+ cell in the crypt-base has been considered the sole intestinal stem cell marker, but a recent study has reported that various epithelial cells of the isthmus region beyond the crypt also have a stemness potential and are involved in intestinal epithelial cell homeostasis [27]. Therefore, studies related to interactions between multiple cell types are needed to understand the mechanisms of action of complex intestinal epithelial cells. Several cell lines can be used as in vitro platforms to study the responses of intestinal epithelial cells. The inflammatory response regulatory function of short-chain fatty acids, including acetate, propionate, and butyrate, was evaluated in Caco-2 cells, a widely used human intestinal epithelial cell line [28]. Caco-2 cells were treated with 5-Fluorouracil, which is used as a chemotherapy drug for cancer, but caused intestinal mucositis as a side effect, to induce intestinal inflammation. Yue et al. reported that three kinds of short-chain fatty acids inhibit the activation of NLRP3 inflammatory bodies (caspase-1, IL-1β, and IL-18) and increase the expression of gut barrier integrity indicators (Occludin and MUC2) compared with inflammation-induced Caco-2 cells. The use of two human enteroendocrine cell lines, NCL-h716 and HuTu-80, has been reported in studies related to hormone responses, another epithelial cell function [29]. Larraufie et al. reported that short-chain fatty acids (especially propionate and butyrate) strongly regulate hormone production in vitro. The use of epithelial cell lines as an in vitro platform has advantages, such as high reproducibility and economic efficiency. Available epithelial cell lines from domestic animals are very limited thus, the most of domestic animal epithelial cell studies done by using IPEC-J2. For example, probiotic L. reuteri, which is isolated from healthy piglet, modulated intestinal health-related factors in LPS-challenged IPEC-J2 cells [30]. Under the challenge conditions, the expression of inflammatory cytokines (TNF-α and IL-6) increased and the expression of tight junction proteins (Claudin-1, Occludin, and ZO-1) expression decreased. However, the above indicators were restored to normal levels through treatment of L. reuteri culture supernatant to IPEC-JC2 cells. However, there are some limitations to using these cell lines in in vitro assays. For example, because most intestinal epithelial cell lines have only a few epithelial cell types, it is difficult to reproduce the epithelial cell combination of complex gut tissues. In addition, immortalization is required to make a stable cell line that can affect the biological function of cell types [31]. Therefore, a better in vitro platform is essential to understand the function of intestinal epithelial cells, and for this reason, intestinal organoids have recently attracted attention [32,33].
Intestinal organoids have also been established in domestic animals and their applications have been reported in various studies, including physiological gut function, immunity, and nutrition [34,35]. For example, pig organoids have been used to investigate the mechanism by which deoxynivalenol, a major mycotoxin, inhibits gut epithelial cell development [36]. Deoxynivalenol does not affect the formation efficiency of organoids, but it reduces the differentiation efficiency of organoids and proliferation of epithelial cells by suppressing the Wnt/β-catenin pathway. Domestic animal intestinal organoids can also be used to evaluate immunomodulatory effects of feed additives. A recent study on the immune response to feed additives containing organic acids and essential oils in S. enterica challenged chicken organoid model was reported [37]. Treatment with feed additives reduced the bacterial load on organoids and downregulated inflammatory responses by decreasing the gene expression of cytokines and chemokines associated with innate immunity. For intestinal epithelial cell studies, the IPEC-J2 cell line, which is isolated from the neonatal piglet mid-jejunum and is not transformed, has been used in various research fields and has provided important insights into dynamic gut physiology [38]. Although the IPEC-J2 cell line is a useful tool for investigating pig intestines, it has several limitations. For example, unlike other epithelial cell lines, IPEC-J2 cells present a high level of transepithelial resistance, a key parameter of epithelial tightness that affects intracellular processes [39,40]. Additionally, a comparison of IPEC-J2 cells and actual pig jejunum tissue under common culture conditions (using fetal bovine serum) revealed differences in ion transport properties and cell morphology [38]. Recently, a transcriptome analysis of IPEC-J2 cells, jejunal organoid, and primary gut tissue was reported [41]. A comparison of gene profiling specifically expressed in pig small intestine with primary tissue and in vitro systems showed a pattern of gene expression similar to that of primary tissue in jejunal organoids rather than IPEC-J2 cells. In addition, there was a clear difference in epithelial cell type marker gene expression between the gut tissue and the IPEC-J2 cell line, and the degree of intestinal epithelial cell marker gene expression in primary gut tissue in organoids was similar. The results of this study suggest that it may be more realistic to use organoid models than cell line-based in vitro platforms to study intestinal epithelial cell regulation in pigs. In our study, MUC2 (for goblet cells) and CHGA (for enteroendocrine cells) were compared based on gene and protein expression level (by immunofluorescence), and it was confirmed that they were not expressed in IPEC-J2 cells but expressed in intestinal organoids. In addition, LYZ (for Paneth cells) and ALPI (for enterocytes) were highly expressed in pig intestinal organoids compared with IPEC-J2 cells. Our data suggested that 3D organoids are better in vitro systems as they have essential epithelial cell types compared with IPEC-J2 cells. In future studies, comparative research on the functions of epithelial cells should be conducted to determine the advantages of pig intestinal organoids.
The intestine has various physiological functions, such as immune regulation, physical barriers to harmful external materials, and nutrient absorption [25]. Nutrient absorption is mainly mediated by enteroendocrine cells and enterocytes in intestinal epithelial cells. Enteroendocrine cells and enterocytes are more directly related to nutrition than other epithelial cell types. Enteroendocrine cells are distributed throughout the epithelium of the gastrointestinal tract, including the small intestine. They release several gut hormones in response to food intake and control gut motility or other endocrinal response [42]. Enterocytes are the major cells responsible for nutrient absorption and are the highest proportion of epithelial cell types [43]. Dietary food delivered to the intestine lumen is broken down into small units of nutrients by digestive enzymes that are sensed by enteroendocrine cells or enterocytes. Enteroendocrine cells secrete gastrointestinal hormones that regulate various organ systems, and enterocytes directly absorb small nutrients [44]. A sequential process involving intestinal digestive enzymes, nutrient-specific transporters, and hormone secretion is required for this series of nutrient-related processes. The intestinal organoid system has three characteristic factors and functional response of nutrient use [17,45]. For example, there was a difference in the degree of nutrient absorption when nutrient-related transporters were treated with transporter inhibitors or transporter gene knock-out organoids, and there was also a difference in the levels of secreted hormones in hormone gene knock-out organoids [45]. These results suggest that the gut organoids play a functional role in nutrient processing. However, there is no clear information regarding the nutrient process-related functions of 3D organoids compared with those of IPEC-J2 cells. In our study, we found that 3D organoids differed in the expression of CHGA and ALPI associated with the nutrient process compared with the IPEC-J2 cell line (CHGA: expressed only in 3D organoids; ALPI: higher gene expression in 3D organoids). Some nutrient process-related gene expression also showed similar differences between 3D organoids and the IPEC-J2 cell line (Supplementary Fig. S1).
Although organoid systems have several advantages, they have structural limitations related to the physical properties of the intestine. The apical side of epithelial cells, which are in contact with the lumen of the intestine and consist of microvilli, performs various biological functions, such as mucus secretion, gut microbiota sensing, and nutrient processing [46]. 3D intestinal organoids, a general culture method, are not exposed to the outside of the intestinal lumen; therefore, some studies require advanced techniques such as microinjection [47,48]. Owing to the 3D organoid structure, several organoid culture models have been reported to modify the 3D culture method according to the purpose of the study [20,49–51]. For example, when comparing physical indicators, such as permeability measurements, the structure of organoids affects the outcome of study. Some receptors that regulate the physical function of intestinal epithelium exist in the apical membrane of epithelial cells. Thus, 3D organoid may not suitable model to understand regulatory response of gut permeability induced by receptors [52]. In this study, we developed a pig 2D organoid model and compared it with a pig 3D organoid model, focusing on nutritional perspectives. Surprisingly, the 2D organoid model had CHGA and ALPI, which are related to nutrient processing, and when fully developed, their expression levels are significantly higher than those of the 3D organoid model. Furthermore, the expression of most nutrient transporter genes was higher in 2D pig organoids. In line with this, many gastrointestinal hormones and digestive enzyme genes were highly expressed on the pig 2D organoid model. These results suggest that pig 2D intestinal organoid models not only have structural and characteristic advantages in nutrition-related studies, but also have superior potential for nutritional physiological responses.
As gene expression levels have limited information, we further attempted to confirm the functional activity of nutrient processing and related response by comparing 3D and 2D pig organoids. We compared the nutrient absorption of two pig organoid models using a fluorescent fatty acid analog. Fluorescent-conjugated nutrients are widely used in studies of nutrient uptake in organoids for intestinal function research [53,54]. Basic organoid cultures have an apical membrane formed inward, similar to the actual intestinal shape, and these structures may affect uptake in the intestinal lumen. Therefore, studies on physiological phenomena such as nutrient uptake and drug absorption have reported changes in the culture methods of intestinal organoids [54,55]. In previous study, differences in nutrient uptake according to organoid culture method have been reported [56]. An apical-out culture method, which exposed the apical side of the intestinal epithelium that absorbs nutrients from the outside, absorbed more nutrients (including fatty acid, amino acid, and glucose) than organoids of the conventional method. Thus, structural property of organoid models should be considered for conducting nutritional study. Compared with the pig 3D organoid model, we found that the pig 2D organoid model absorbed more fatty acids during the same period. Although the exposed apical membrane of the 2D intestinal organoid model facilitate the absorption of fatty acids, nutrients can be diffused or actively transported depending on their type, and the degree of uptake can vary depending on the location of nutrient transporters (basal, apical, or both) [57]. Therefore, suitable culture models for target nutrients should be considered in future organoid-based nutritional studies.
Fatty acids, one of the major nutrients, are broken down from dietary lipids in the intestine and absorbed mainly by enterocytes. Fatty acid metabolism involves several proteins including receptors, transporters, and binding proteins [58]. Unlike other nutrients, fatty acid absorption pathways exhibit unique properties. First, fatty acids are re-esterified by related complex molecules before entering enterocytes through the apical membrane. The absorbed fatty acids are then packaged into pre-chylomicrons or stored as intracellular lipid droplets for fatty acid oxidation. Finally, mature chylomicrons are released from the enterocytes and transported throughout the body via the lymphatic system [59]. Nutrients can induce various signals in the gastrointestinal tract, including peptide hormone release by enteroendocrine cells. These hormones act efficiently over the short term and are secreted by several types of enteroendocrine cells that secrete hormones such as glucagon-like peptide 1 and 2 (GLP-1 and GLP-2), cholecystokinin (CCK), glucose-dependent insulinotropic polypeptide (GIP), and peptide YY (PYY) [42]. As an example of hormone release by fatty acids, it has been reported that LCFAs significantly induce the release of GLP-1 and GLP-2 in the pig gut tissue ex vivo culture model [60]. To confirm the response to fatty acids, especially LCFAs, in pig intestinal organoid models, we compared the gene expression levels of fatty acid metabolism-related proteins and hormones after treatment with LM. In summary, no significant differences were found in the 3D organoid model; however, in the 2D organoid model, the LCFA receptor and several hormone-encoding genes showed an overall increase after LM treatment. FFAR1 and FFAR4 (as well-known GPR 40 and GPR120, respectively) are representative LCFA receptors as types of G protein-coupled receptor (GPCR). And they that are expressed in several enteroendocrine cell types and are associated with hormones [61]. Some gut hormones, including CCK, GLP-1, and PYY, are well known for their anorexic activity, in which their concentrations rise soon after food ingestion and remain elevated for up to several hours, depending on meal size and composition [62]. Lipid intake in food stimulates the release of these hormones and increases their plasma concentrations [63,64]. To support the association between receptors and hormones, deficient mouse models of FFAR1 or FFAR4 have been shown to impair lipid-induced hormone secretion responses [65,66]. Our 2D pig intestinal organoid model suggested that fatty acids can be recognized by receptors in the apical membrane and can simulate hormonal responses similar to those in the intestinal lumen. However, since there are limitations due to comparisons at the genetic level, it is necessary to directly compare hormone secretion levels or study the mechanism of the intracellular fatty acid-induced pathway up to hormone release.
LCFAs can be divided into saturated and unsaturated fatty acids based on their 12–20 carbon chain composition. LCFAs absorbed by cells are regulated by several metabolic responses, including cellular metabolism, energy homeostasis, and cell proliferation [67]. Various LCFAs, like other nutrients, can diffuse or be transferred through specific proteins. First, LCFAs uptake is carried out by plasma membrane-associated fatty acid-binding protein (FABPpm) such as fatty acid transport protein 4 (FATP4), and cluster of differentiation 36 (CD36) in enterocytes. LCFAs absorbed by acyl-CoA synthetase (ASC) are present as free fatty acids or fatty acyl coenzyme A (fatty acyl-CoA). They are then bound by fatty acid-binding protein (FABP) and acyl-CoA binding protein (ACBP) and trafficked into the cells [68]. Fatty acyl-CoA migrates to the mitochondria, and intramitochondrial oxidation proceeds via the beta oxidation pathway. The mitochondrial matrix does not contain enzymes that activate fatty acids containing 14 or more carbon atoms. Thus, the entry of LCFAs into the mitochondria is regulated by specific enzyme activities, such as carnitine palmitoyltransferase 1 (CPT 1) and CPT 2. Acetyl coenzyme A (acetyl-CoA) is produced as an end product of beta oxidation, and it promotes ATP synthesis through the tricarboxylic acid (TCA) cycle [69]. To confirm LCFAs-induced metabolism in pig organoid models, mitochondria staining and intracellular ADP:ATP ratios were compared. In our study, LM treatment increased mitochondrial fluorescence intensity in 2D organoids but not in 3D organoids. In addition, there was no significant difference in the ADP:ATP ratio after LM treatment in 3D organoids, whereas it decreased in 2D organoids. The ADP:ATP ratio reduction may imply the presence of a larger proportion of ATP within the cells, which, together with the mitochondria staining results, may have contributed to the generation of ATP through fatty acid oxidation within the 2D organoids. However, no changes were observed in 3D organoids, which may have caused poor fatty acid transfer into the cells or differences in fatty acid oxidation-related enzyme activity. Collectively, 2D and 3D organoid systems show different physiological response in nutrient metabolism maybe due to poor nutrient absorption and/or lower expression of nutrient process-related cells and gene expression.
CONCLUSION
In summary, our results suggested that pig intestinal organoids are more suitable for intestinal epithelial cell research than the existing in vitro systems such as IPEC-J2. Furthermore, we have established a 2D organoid model for intestinal lumen research and further compared nutrient-related properties, such as nutrient transporters, hormones, and digestive enzymes, with a 3D organoid model to characterize them. Compared with the 3D organoid model, the established 2D organoid model showed more active absorption of nutrients, gene expression, and metabolic processes related to nutrient responses. These findings emphasize the suitability of the 2D organoid model as an in vitro platform for nutrition-related research and provide an improved understanding of nutrient use by intestinal epithelial cells. This study provides essential information for further investigations of the interactions between intestinal epithelial cells and nutrients in the gut environment.
