Lactococcus lactis is a lactic acid bacterium (LAB) that has been designated by the United States Food and Drug Administration as generally recognized as safe. Lactococcus lactis subsp. lactis is frequently present in naturally fermented dairy products and is widely employed in commercial feed, milk fermentation, and vaccine manufacturing [1]. However, several studies have indicated that L. lactis can cause mastitis in cows and it has even been associated with clinical cases (e.g., lactococcosis in silver carp, liver and spleen disease in waterfowl, and endocarditis in humans). A functional genomic study revealed that dairy L. lactis subsp. lactis diverged from plant-associated ancestors independently of human intervention, but was later selected for its functional properties in dairy fermentation [2]. Selection pressure leads to a reduction in the genome and the loss of genes, while acquiring genes involved in protein and lactose metabolism through horizontal gene transfer (HGT) [3]. HGT is also believed to be the main reason for the transmission of genes for antimicrobial resistance (i.e., erythromycin) and virulence factors from other species, conferring harmful traits to L. lactis subsp. lactis [4]. Further efforts are required to fully understand the evolutionary divergence of the group and to better understand the differences between safe and potentially pathogenic strains of L. lactis.
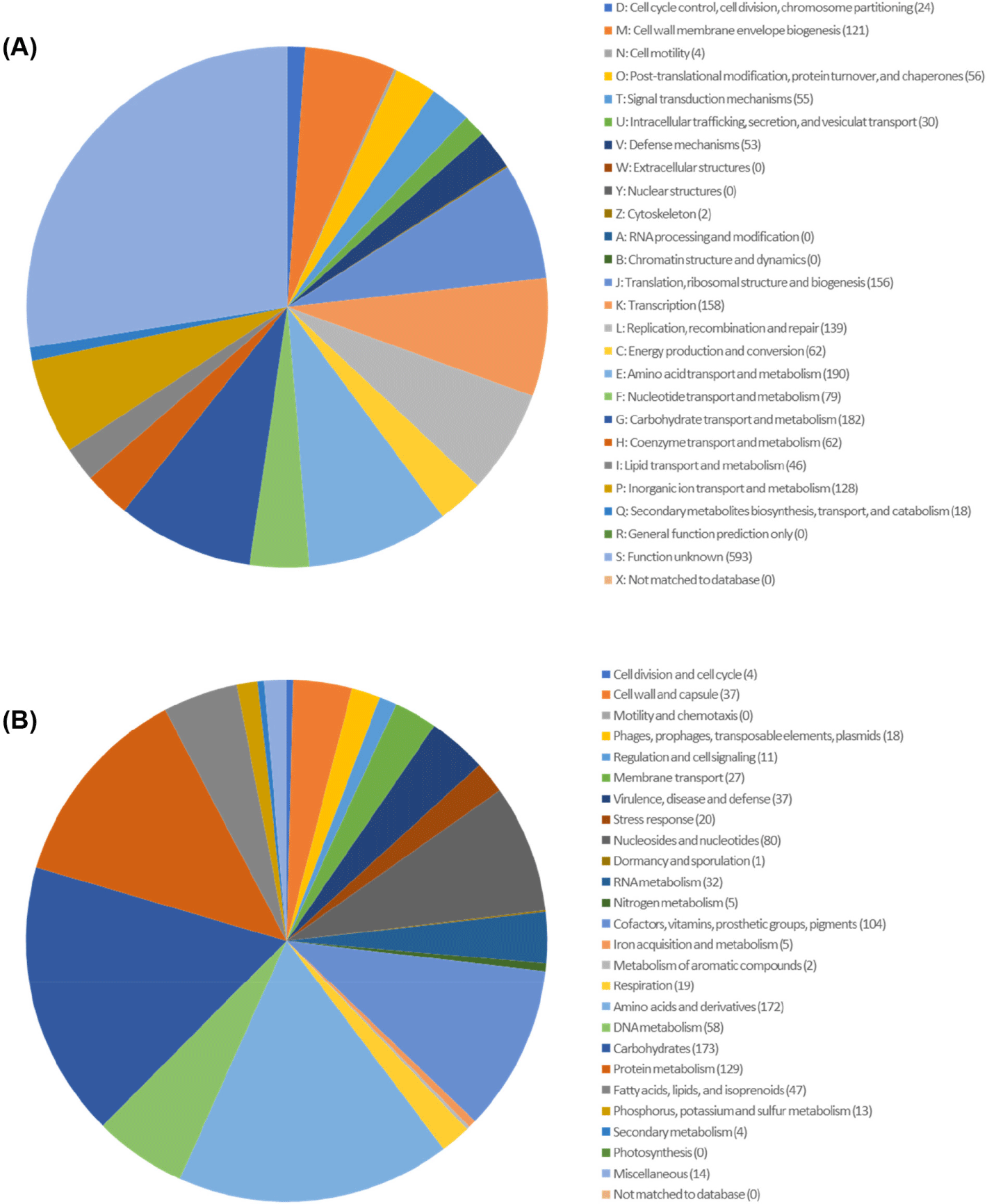
Here, we present the whole genome of Lactococcus lactis subsp. lactis DOME 6301 strain, which has the ability to produce antimicrobial compounds, isolated from raw cow milk. Strain DOME 6301 was routinely cultured in de Man, Rogosa, and Sharpe (BD Difco) broth supplemented with 0.05% L-cysteine HCl (Sigma Aldrich). Genomic DNA was extracted from 12–15 h cultures using the QIAamp PowerFecal DNA Kit (Qiagen) following the prescribed protocol. Sequencing was performed at CJ Bioscience using the Pacific Biosciences RSII Single Molecule Real-Time platform with a 20-kb SMRTbellTM template library (PacBio), followed by de novo assembly of the reads using FALCON 0.5. Whole-genome analysis of L. lactis subsp. lactis DOME 6301 (Fig. 1) revealed a genome of 2,532,858 base pairs with a guanine and cytosine content (G + C content) of 35.0% and an N50 value of 2,417,727 bp, assembled into three contigs, one of which was designated as plasmid pDOME6301-LcnB (103,795 bp). The genome consisted of 2,469 protein-coding genes, 78 tRNA genes, and 22 rRNA genes (Table 1). Genome annotation and functional categorization were performed using Rapid Annotation Subsystem Technology (http://rast.nmpdr.org/) with default parameters, and a cluster of orthologous groups was obtained from the EZBioCloud server. As shown in Fig. 2, most genes were predicted to be involved in cell wall and membrane envelope biogenesis (121); translation, ribosomal structure, and biogenesis (156); amino acid transport and metabolism (190); carbohydrate transport and metabolism (182); and inorganic ion transport and metabolism (128).
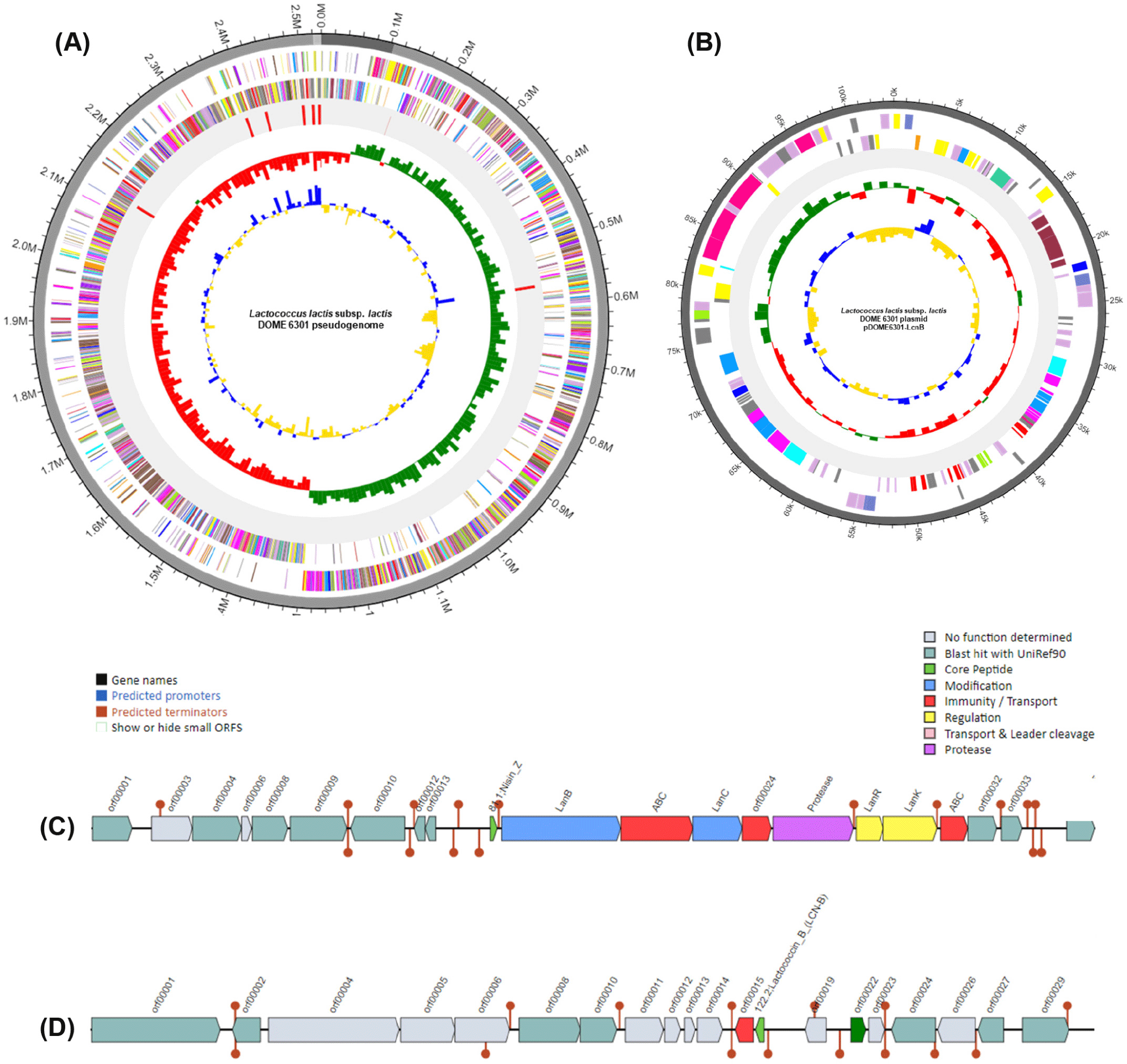
Among the predicted carbohydrate-related genes, several genes encoding enzymes for complex carbohydrate utilization were found, including cellulase, endo-1,4-β-xylanase, oligosaccharide reducing-end xylanase, diamine N-acetyltransferase, α-amylase, cyclomaltodextrinase, pullulanase, non-reducing-end α-L-arabinofuranosidase, and an uncharacterized multiple-sugar transport system permease YteP, which may hold significant function in carbohydrate utilization in the animal host. Additionally, two bacteriocin gene clusters corresponding to nisin Z (Class I; chromosomally encoded) and lactococcin B (Class IID; plasmid-encoded) were identified using the BAGEL4 webserver (http://bagel4.molgenrug.nl/) as depicted in Fig. 1C and D. Downstream the nisin Z open reading frame (ORF) contained genes for bacteriocin modification (lanB and lanC), regulation (lanR and lanK), immunity and transport (nisT, nisF, and nisin immunity proteins), and a serine protease for leader peptide cleavage. The lactococcin B operon contained an ORF for the core peptide and an immunity protein. Preliminary experiments showed that the bacteriocins produced by strain DOME 6301 inhibited the growth of oral pathogens, including Streptococcus mutans KCTC 5365, Prevotella intermedia KCTC 15693T, and Fusobacterium nucleatum KCTC 2488T, implying that this strain could be used as a probiotic candidate for the development of functional dairy products having antimicrobial properties.
On an evolutionary level, the strains are thought to differ based on their carbohydrate metabolism, ability to defend themselves by producing antimicrobial compounds, and how they react to stress [5]. Antimicrobial resistance genes were predicted using the Comprehensive Antibiotic Resistance Database Resistance Gene Identifier [6], which revealed the presence of vanY (% ID, 33.7%) and qacJ (% ID, 46.67%) genes associated with resistance to glycopeptide antibiotics and disinfecting agents or antiseptics, respectively. Furthermore, genes encoding virulence factors, including hemolysin (hemolysin-3, a conserved virulence factor) and enterotoxin were detected in the chromosome (Supplementary Information). Despite the significant potential of strain DOME 6301 in various industrial applications owing to the presence of enzymes for the breakdown of complex carbohydrates, the presence of genes involved in hemolytic activity and enterotoxins might limit its potential use. Nevertheless, nisin Z, a structural variant of the commercially accepted nisin A (His27 > Asn), remains valuable for pathogen control and is a possible alternative to conventional antimicrobials [7]. These observations contribute to the elucidation of the evolutionary background of L. lactis subsp. lactis and highlight the importance of intensive and accurate characterization of LAB strains for their potential use in the fermentation industry or for the development of functional probiotics.
