INTRODUCTION
Goats are widely considered to be one of the earliest species to be domesticated. The bezoar goat (Capra aegagrus), which inhabits the rugged mountainous regions stretching from the Taurus Mountains in Turkey to Pakistan, is considered to be the wild ancestor of the domestic goat (Capra hircus) [1]. It is therefore plausible that the indigenous goat breeds found in present-day Turkey are direct descendants or closely related to the ancestral populations.
Goat breeds are classified into dairy, meat, fiber, and combined productivity breeds. The Angora goat holds significant prominence in Turkey due to its crucial role in mohair production, characterized by its lengthy and glossy coat [2]. Angora goats have been raised in numerous countries for an extended period, including Türkiye, Australia, America, Argentina, New Zealand, France, and South Africa. The origins of this breed can be traced back to Ankara, the capital city of Türkiye. Mohair is a valuable animal product and an important raw material for the textile industry. It absorbs moisture easily, it is durable, shiny, elastic, resistant to harmful sun rays, heat-resistant, highly insulating, and easy to dye [3]. The economic value of fiber is primarily determined by its fineness and quantity, which are considered to be the most crucial factors. The value of mohair in world markets depends primarily on its quality characteristics, including fineness, quantity, length, curl, and strength. Therefore, efforts are being made in mohair-producing countries to enhance these characteristics and improve the overall quality of the product [4].
Before these traits are accepted as selection criteria, genetic parameters need to be estimated. Numerous research studies have consistently shown that the characteristics of hair follicles (HF) and the genetic regulatory mechanisms associated with them have a direct impact on the production and quality of hair fibers [5–16]. Enhancing our knowledge and comprehension of the developmental and biological properties of the fibers generated by HF can offer valuable strategies for obtaining fibers that possess the desired properties [6]. Animal fibers are generated through the HF found in the skin. These fibers possess unique mechanisms that enable their growth, development, and self-renewal. The HF themselves are regarded as miniature organs that arise from the interaction between neuroectodermal and mesodermal tissues [17]. The embryonic period initiates hair follicle formation, leading to its full growth within the same developmental stage [4]. The initial phase of development involves a series of stages collectively known as morphogenesis, including induction, organogenesis, and cytodifferentiation [18]. Following birth, HF undergo a recurrent process known as the hair follicle cycle. This cycle consists of three specific phases: anagen, catagen, and telogen. Each phase of the hair follicle cycle has a unique pattern of gene activation and suppression. The transition between stages is intricately controlled by transcription factors and enzymes, which are influenced by specific signaling molecules present in the immediate environment. These signaling molecules consist of cytokines, hormones, neurotransmitters, and essential mediator molecules [19].
During the anagen phase, HF undergo a period of rapid growth. Subsequently, in the catagen phase, the hair bulbs undergo contraction, resulting in a gradual thinning as they ascend. HF in the telogen phase are situated solely above the sebaceous glands and remain in a stationary condition [20]. The cycle initiates as the HF reaches maturity and enters the catagen phase in response to various activating signals. Apoptosis is a distinctive feature of the catagen phase, wherein programmed cell death takes place in the inner root sheath (IRS), outer root sheath (ORS), and hair matrix. This apoptotic process leads to the regression of the lower two-thirds of the hair follicle, ultimately resulting in the development of the epithelial column. Simultaneously, the dermal papilla (DP) ascends, approaching the bulge region in close proximity. During the telogen phase, the main hair shaft (HS) experiences a change and transforms into club hair, all the while maintaining the close proximity of the DP to the bulge. The onset of the anagen phase is triggered by the interaction between the bulge and the DP. During this stage, the activation of hair follicle stem cells occurs in the upper segment of the hair follicle. Subsequently, the hair follicle starts to grow downward, giving rise to the bulb and other related structures. As a result, new hair is produced while the old club hair is shed [21]. These phases, the duration of which varies from organism to organism and from follicle to follicle, are the anagen, catagen, and telogen phases, respectively. In goats, the secondary follicles that form cashmere and mohair fibers enter the active anagen phase between June and November and grow for about 185 days. This is followed by the catagen phase, which lasts about 60 days between December and January. From February to the end of May, the telogen phase extends for approximately 120 days [22].
Hair follicle regulation involves the activation of specific genes known as Keratin genes (KRTs) and the presence of Keratin-associated proteins (KAPs). Moreover, the growth and maturation of HF are profoundly impacted by a multitude of signaling pathways, including but not limited to MAPK, PI3K-Akt signaling, Ras signaling, and the cell cycle [20]. Nevertheless, the exact features, chronological order, and intersections of these signaling pathways are yet to be fully elucidated.
To produce finer animal fiber, it is crucial to comprehend the regulatory role that genes, signaling pathways, and other processes play in hair follicle growth. Transcriptome sequencing technology was utilized in this study to explore the mRNA expression profile of goat skin tissue. The research was conducted on two Ankara goat varieties, Eskisehir and Lalahan, with both males and females studied in three different growth phases: anagen, catagen, and telogen. The aim of this study was to determine the mRNA expression profile of skin tissues collected from Angora goat varieties using RNA-seq analysis. This is the first study to use the mRNA expression profile of skin tissues to analyze differences between Angora goat varieties in the anagen, catagen, and telogen phases for both males and females.
MATERIALS AND METHODS
The animal material for this study consisted of Lalahan and Eskisehir varieties of Ankara goats. The Lalahan variety of Ankara goats has been purebred since 1930 and has been under protection since 1997. The Eskisehir variety of Ankara goats carries US Ankara goat blood in its genotype and was brought to the International Center for Livestock Research and Training (ICLRT) in 1997 from the Eskisehir Anatolian Agricultural Enterprise as part of the “Protection of Animal Genetic Resources” project conducted by TAGEM, which is the General Directorate of Agricultural Research and Policies under the Ministry of Agriculture and Forestry. For this study, four female and three male Lalahan variety Angora goats and three female and three male Eskisehir variety Angora goats (located at 39°58’16.71”N latitude and 33°6’33.27”D coordinates) were used. Angora goats in each group were selected from clinically healthy goats born in 2021, and the same management and nutritional conditions were applied to all animals.
Three skin samples were taken from each goat as biological duplicates at every stage of hair follicle growth (September, January, and March). In total, 72 samples were analyzed in this study, including samples from two Angora goat varieties, two gender, three developmental stages (three biological replicates), and two technical replicates for RNA sequencing.
Skin tissue samples were taken from the right shoulder region, which is the upper one-third of the left scapula, of these goats during the anagen phase (September), catagen phase (January), and telogen phase (March) using a 0.3 mm diameter punch biopsy tool while adhering to sepsis and antisepsis protocols. The shoulder region was shaved and wiped with 70% alcohol prior to the biopsy. For each animal, prilocaine hydrochloride was applied subcutaneously for local anesthesia to reduce animal pain. A 0.3 mm diameter punch biopsy instrument was used to obtain a biopsy sample from the shoulder region, and the biopsy materials were placed in an RNA preservation solution to prevent degradation and stored at –80°C until RNA isolation. Ethics committee permission dated September 15, 2022, and numbered 205 was obtained from the ICLRT Animal Experiments Local Ethics Committee for this study.
The extraction of RNA from the skin tissue samples was carried out utilizing the Qiagen RNeasy® Fibrous Tissue Mini Kit (Qiagen) in accordance with the provided kit protocol. The assessment of RNA quality and quantity was conducted using Thermo NanoDrop 2000 spectrophotometry, measuring the absorbance at A260/Α280 nm wavelengths. RNA purity was determined by ensuring that the absorbance ratio at A260/A280 nm was equal to or greater than 2.0. To assess the integrity and quality of the RNA sample, the Qubit RNA IQ Assay was performed on a Qubit 4.0 fluorometer (Life Technologies) using the Qubit RNA BR Assay Kit (500 Assays, Cat. No: Q10211. Thermo Fisher Scientific). The RNA integrity number (RIN) was evaluated using the Bioanalyzer 2100 system (Agilent Technologies) with the RNA Nano 6000 Assay Kit. For the purpose of RNA sequencing, only RNA samples meeting the criteria of having a RIN value above 8.0 and an OD260/OD280 ratio higher than 1.9 were utilized. The total RNA was appropriately stored in a freezer maintained at a temperature of –80°C.
The cDNA library was constructed using the Illumina Stranded Total RNA Prep Ligation with Ribo-Zero Plus kit, adhering to the guidelines provided by the manufacturer. The Illumina Ribo-Zero Plus rRNA Depletion Kit was used to deplete rRNA from the samples and to obtain mRNA before RNA sequencing. The Illumina stranded total RNA kit was used to generate libraries from ribosomal RNA-depleted samples. The constructed cDNA library was sequenced using the Illumina Nova Seq 6000 platform, and paired-end reads of 150 base pairs (bp) were generated during the sequencing procedure.
Within the scope of the study, expression counts of goat (Capra hircus) samples were calculated, and comparisons were performed between the groups. The quality of the raw data was evaluated using the FastQC software. The raw data in FASTQ format underwent primary quality control as the first step in the processing pipeline. In this stage, raw reads including more than three poly-N sequences or adaptor sequences, as well as those containing a base percentage with a quality value, were filtered away in order to produce clean reads. All further analyses were performed using only the high-quality, clean data from the previous processing step. Then, the Trimmomatic tool (version 0.39) was used (i) to trim the reads with a phred score less than 20 and (ii) to filter out the reads shorter than 20 bases long. Moreover, Kallisto v0.44.0 was utilized to estimate transcript abundance by pseudo aligning the reads [23].
The expression values at the gene level were obtained using the Kallisto tool (version 0.46.1) and the tximport R package. Capra hircus cDNA, non-coding RNA (ncRNA), and gtf files were obtained from the genome database [24]. cDNA and ncRNA were combined and indexed using the Kallisto tool. Transcript-level raw counts were obtained using the Kallisto quant function. The computation of gene-level raw counts was carried out using the tximport tool. The alignment of clean reads from each sample to the goat reference genome, ARS1.2 (GCA_001704415.2) [25], was performed using the HISAT2 software. StringTie software was employed for the tasks of transcriptome assembly, annotation, and expression calculations [26].
The Detection of DEGs was performed by the DESeq R package [27]. DESeq first normalizes raw counts and uses the normalized values for statistical analysis. Prior to normalization with DESeq, genes with a count of zero were removed from the calculation in all the groups being compared. Principal component analysis (PCA) was performed using all samples to test for the presence of outliers. The data used for PCA analysis was the DESeq normalized gene counts. According to this result, no outlier sample was detected, and differential expression analysis was conducted using all samples. The adjusted p-values (Benjamini-Hochberg corrected) and fold changes for each gene in the dataset were computed using the DeSeq package.
DEGs were detected by applying a false discovery rate (FDR) threshold of ≤0.001 and an absolute log2Ratio value of ≥1 (representing a two-fold change) as the thresholds for significance. In cases where the fold change exceeded 1 and the FDR was less than 0.05, the observed change was classified as significant upregulation. Conversely, if the fold change was less than 1 and the FDR was less than 0.05, the change was designated as significant downregulation.
In the scope of the differential expression analysis, comparisons were performed for a total of 3 main groups, which include “Gender”, “Variety”, and “Inter Variety” groups. DEGs were identified in the gender group by comparing male and female individuals within each variety at the anagen, catagen, and telogen phases. Therefore, a total of six comparisons were made within this group. In the Variety group, differences in gene expression patterns between anagen and catagen, anagen and telogen, and catagen and telogen phases were analyzed for each variety and gender. Thus, a total of 12 comparisons were performed within this group. Finally, in the inter-variety group, each phase was compared between varieties separately for male and female individuals, and as a result DEGs in each phase were identified between varieties separately in male and female individuals. This group was also subject to a total of six comparisons.
To assess the influence of gender on DEGs associated with hair growth, six distinct groups were defined for each variety: (1) Eskisehir Anagen Female versus Eskisehir Anagen Male; (2) Eskisehir Catagen Female versus Eskisehir Catagen Male; (3) Eskisehir Telogen Female versus Eskisehir Telogen Male; (4) Lalahan Anagen Female versus Lalahan Anagen Male; (5) Lalahan Catagen Female versus Lalahan Catagen Male; (6) Lalahan Telogen Female versus Lalahan Telogen Male.
In the variety group, to identify DEGs between phases in each variety and each hair growth, phases were compared separately in males and females. A total of 12 comparisons were performed within this group. For the purpose of comparing experimental groups, the following contrast groups were established: (1) Eskisehir Female Anagen versus Eskisehir Female Catagen; (2) Eskisehir Female Anagen versus Eskisehir Female Telogen; (3) Eskisehir Female Catagen versus Eskisehir Female Telogen; (4) Eskisehir Male Anagen versus Eskisehir Male Catagen; (5) Eskisehir Male Anagen versus Eskisehir Male Telogen; (6) Eskisehir Male Catagen versus Eskisehir Male Telogen; (7) Lalahan Female Anagen versus Lalahan Female Catagen; (8) Lalahan Female Anagen versus Lalahan Female Telogen; (9) Lalahan Female Catagen versus Lalahan Female Telogen; (10) Lalahan Male Anagen versus Lalahan Male Catagen; (11) Lalahan Male Anagen versus Lalahan Male Telogen; (12) Lalahan Male Catagen versus Lalahan Male Telogen.
The genes that were expressed differentially were subject to enrichment analyses through the use of the g: Profiler tool for investigating their roles in the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) pathways [28] and KOBAS [29] online analysis database. The KOBAS analysis has been conducted by configuring the query parameters to align with the chosen organism, Capra hircus. Subsequent analysis focused on GO terms and KEGG pathways, with significance determined by an adjusted p-value below 0.05. Functional annotations for DE transcripts were performed using the GO database, which assigned them to one of three categories: biological processes (BPs), molecular function (MF), or cellular component (CC). The candidate genes were analyzed using the STRING database [30] to generate a functional protein-protein interactions network, which was visualized with the Cytoscape program [31].
bioDBnet is a tool that allows users to perform ortholog conversions, converting identifiers from one species to those of another species. The conservation of gene identities across species has been analyzed using bioDBnet [32].
To confirm the differential gene expression determined by RNA sequencing, Quantitative Real-Time polymerase chain reaction (qPCR) validation was performed with five randomly selected genes (KRTAP8.1, FOS, FGF5, FOSL1, and FOSB) that exhibited differential expression across the three developmental stages. GAPDH was utilized as the endogenous control in this experiment. The qPCR reaction was conducted using a total volume of 10 µL, comprising of 2 µL of cDNA, 5 µL of SYBR green real-time master mix, and 0.5 µL of each primer. Relative gene expression levels were determined using the 2-ΔΔCT method [33]. At three distinct developmental stages, total RNA extraction was performed on all experimental goat skin samples. The QuantiTect Reverse Transcription Kit (Qiagen, Cat ID: 205311) was used for synthesizing cDNA from the RNA in accordance with the manufacturer’s instructions. The primer sequences are provided in Table S1. The qPCR was conducted on the Bio-Rad CFX96 Real-Time PCR System using the Bio-Rad Universal SYBR® Green Supermix. The accuracy of the SYBR green PCR signal was verified through an analysis of the melting curve, ensuring its specificity.
RESULTS
From 36 samples, a cumulative RNA-Seq dataset of 211.94 GB was generated, with Q20 percentages exceeding 99.69% and Q30 percentages surpassing 99.17% for each sample. The GC content ranged from 42% to 50%. Supplementary File 1 provides a comprehensive overview of quality control and alignment statistics, affirming the high quality and uniformity of the sequencing data across all sample sets. The quality control outcomes ascertain that the sequencing results are robust and reliable, establishing a solid foundation for subsequent data processing.
Following the mapping of clean reads to the goat reference genome, comparisons were made between each growth cycle. Differences in gene expression patterns between anagen and catagen, anagen and telogen, and catagen and telogen phases were compared between varieties and between genders to identify DEGs of hair follicle development and cycle at different stages.
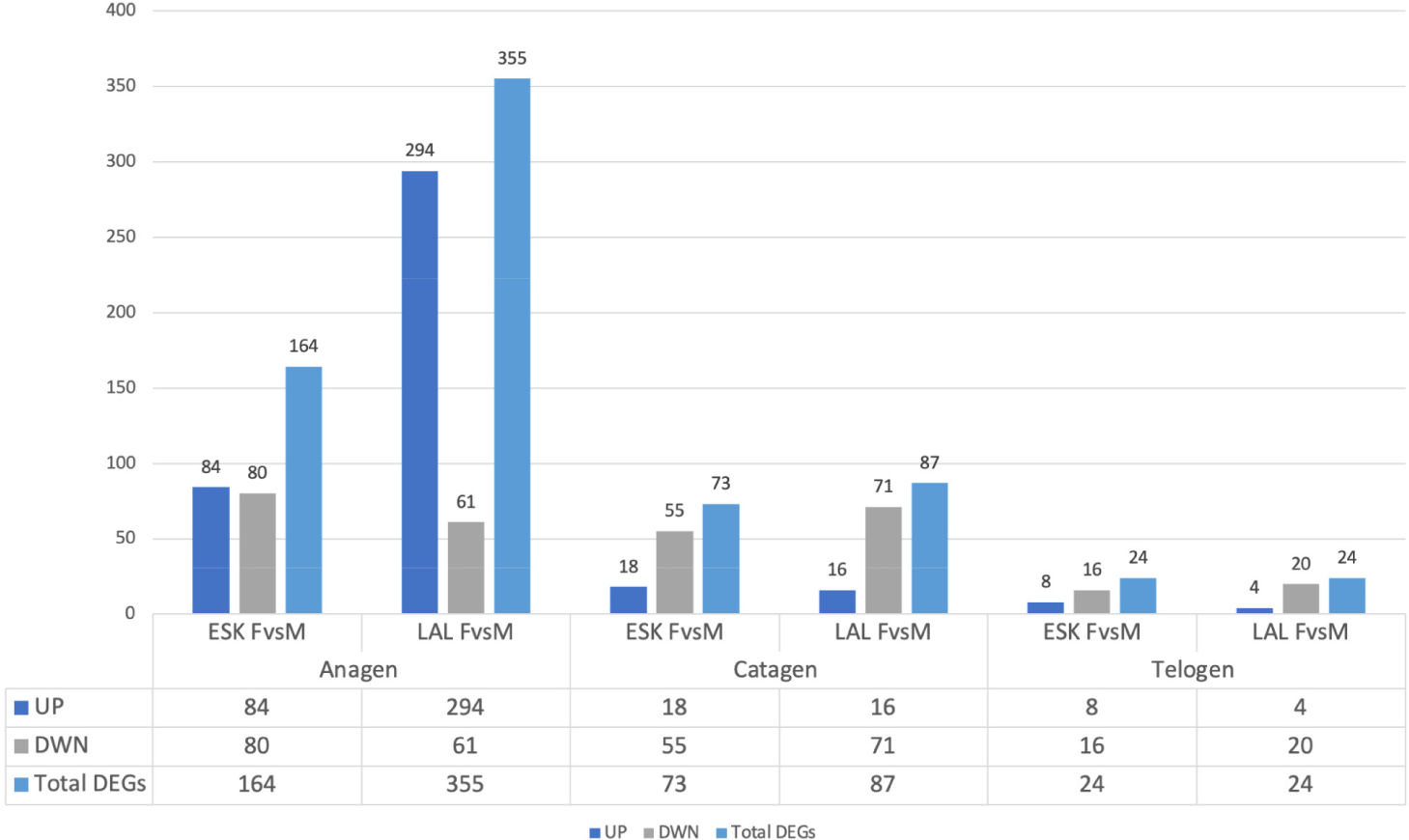
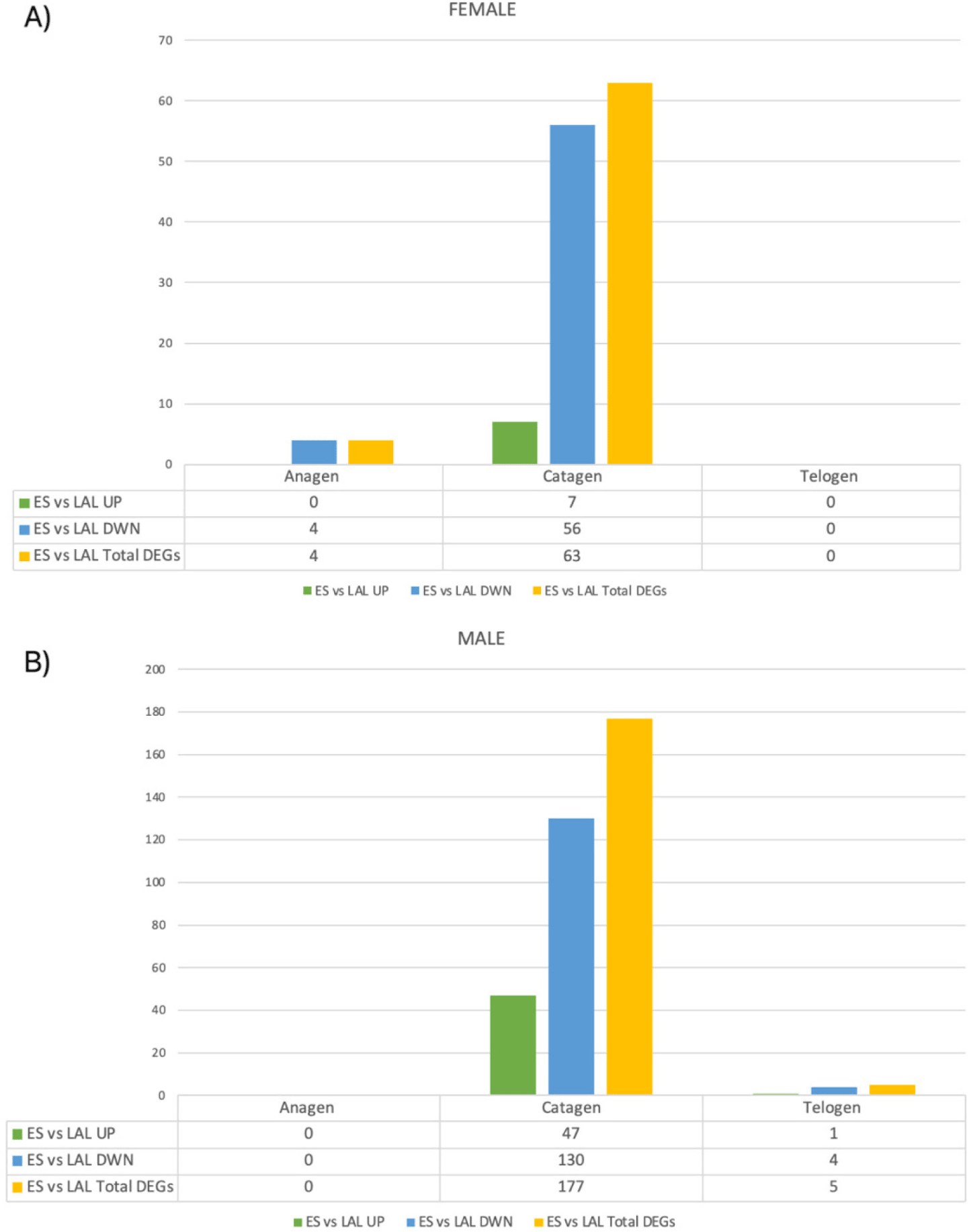
Through the comparison of gene expression profiles in female and male goats within the same variety and hair growth phase, we identified genes exhibiting differential expression between males and females. In the anagen phase, a total of 164 and 355 DEGs were detected between male and female individuals of the Eskisehir and Lalahan varieties, respectively. Furthermore, a notably higher count of DEGs was observed during the anagen phase in the Lalahan Angora goat in comparison to the Eskisehir Angora goat. This suggests that there may be some important differences in the way that these two varieties of goats regulate hair growth. The volcano plot generated for this group is shown in Fig. 1.
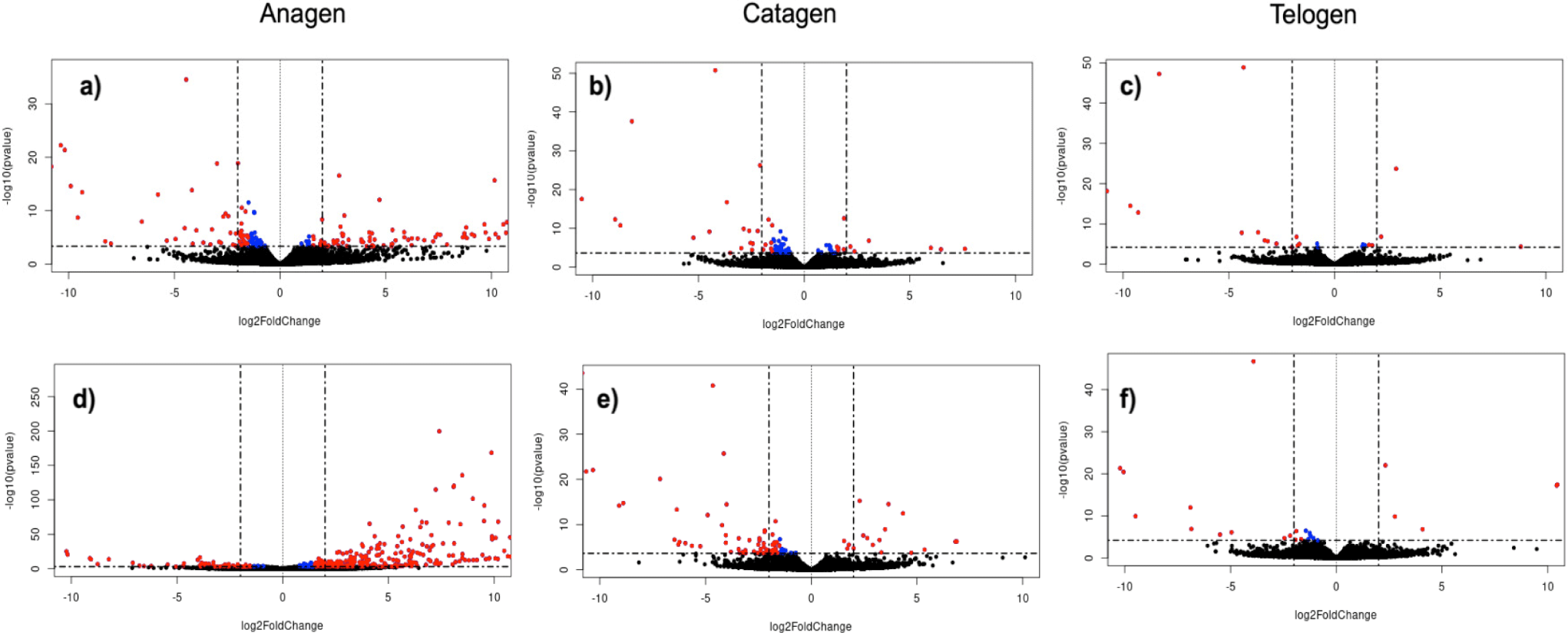
In the catagen phase, a total of 73 and 87 DEGs were detected in the comparison between male and female individuals of the Eskisehir and Lalahan varieties, respectively. Additionally, 24 DEGs were detected in the telogen phase between male and female individuals of both the Eskisehir and Lalahan Angora goats (Fig. 2 and Supplementary File 2). This observation suggests that when comparing male and female individuals in both Eskisehir and Lalahan Angora goats, the differentiation in genes expressed during the anagen phase is more prominent than during the catagen and telogen phases. This information could be used to improve the quality of Angora goat mohair and could be important for understanding the genetic basis of hair growth.
By conducting this comparison, we were able to identify genes that exhibited differential expression among the three different stages within the same gender and variety. Between the anagen and catagen phases in the female Eskisehir variety of Angora goats, a total of 111 DEGs were identified. Among these genes, 30 were upregulated, while 81 were downregulated. On the other hand, a total of 151 DEGs were detected between the anagen and catagen phases in the male Eskisehir variety of Angora goat, of which 100 genes were upregulated and 51 genes were downregulated. Moreover, a total of 18 DEGs were detected between the anagen and catagen phases in the female Lalahan variety of Angora goats, of which 11 were upregulated and 7 were downregulated. Furthermore, a total of 76 DEGs were detected between anagen and telogen phases in the female Eskisehir variety of Angora goat, of which 13 were upregulated and 63 were downregulated. In this group, KRTAP7-1 and KRTAP8-1 were identified as the first two DEGs (padj= 6,09E-15 and 7,04E-14, respectively; and FC:0,2745; 0,2168, respectively. Supplementary File 3). The volcano plot generated for this group is shown in Fig. 3.
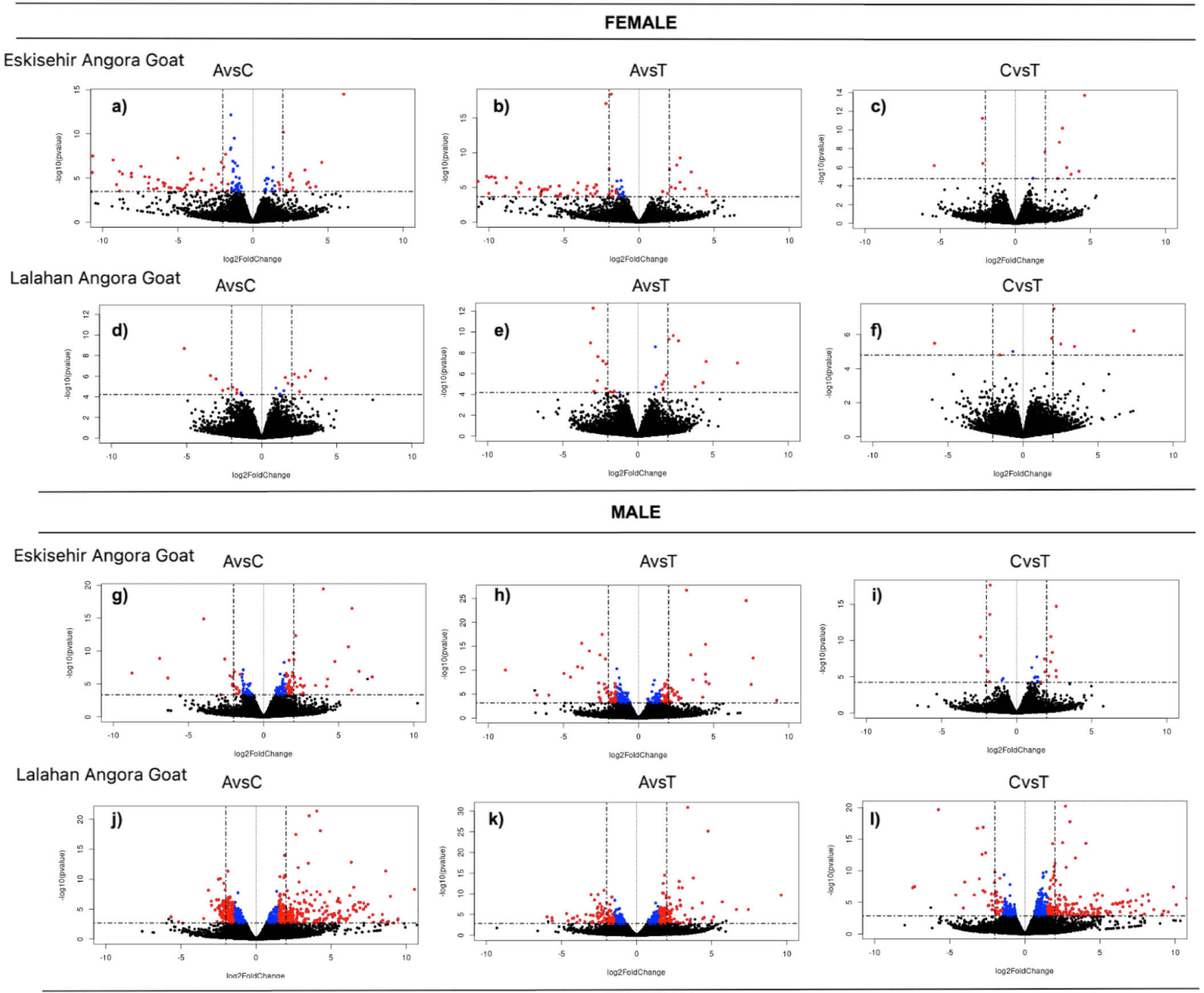
For the female Lalahan variety of Angora goat, a total of 23 DEGs were detected between the anagen and telogen phases, of which 12 were upregulated and 11 were downregulated. In this group, KRTAP7-1 and PTGS2 were identified as the first two DEGs (padj= 9,83E-09 and 2,08E-06, respectively; and FC: 0,127; 5,115, respectively). The comparison results obtained for the other groups are shown in Fig. 4. Gene expression patterns in the male groups appeared to be significantly different from those in the female groups at each hair growth phase.
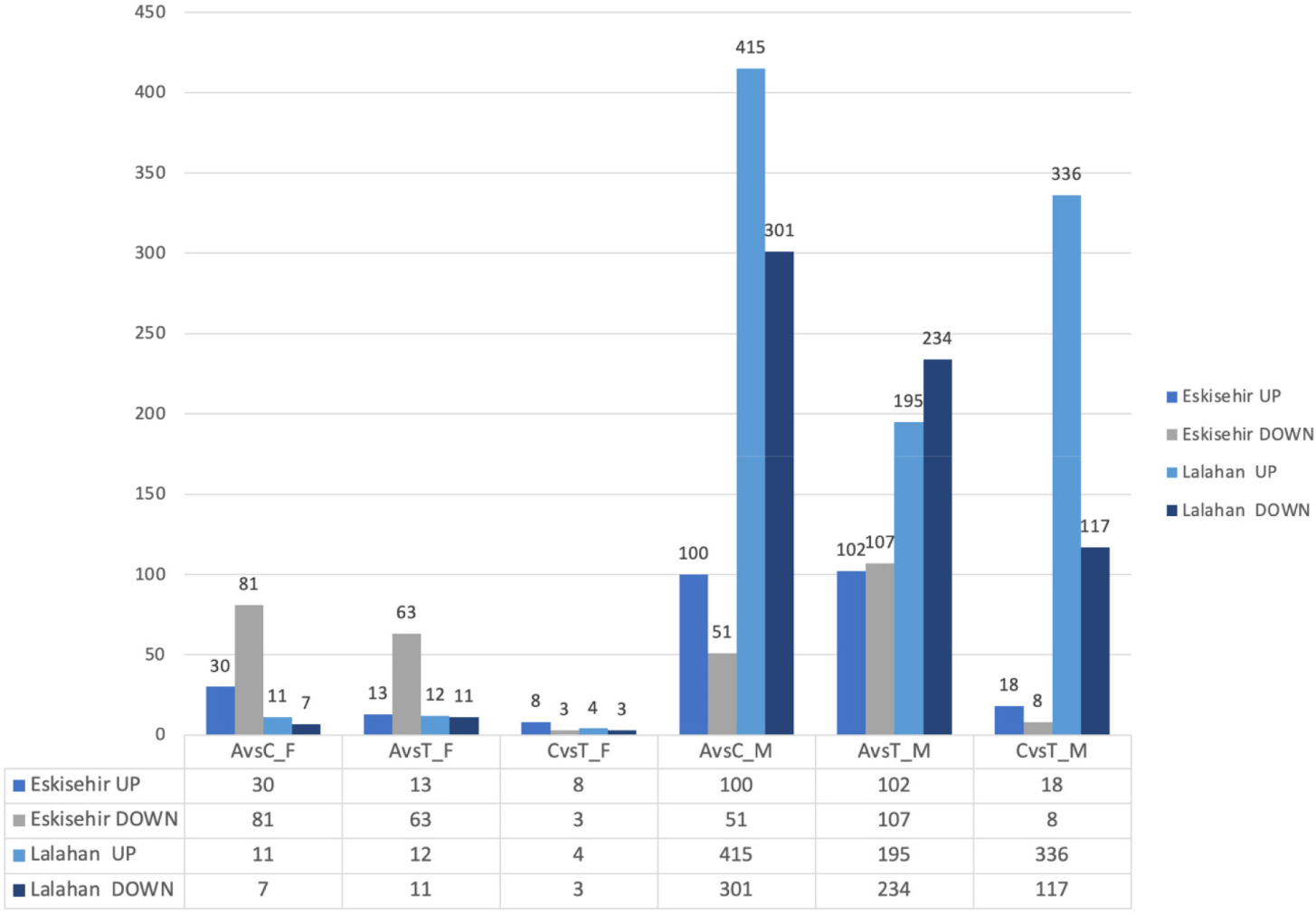
In the inter-variety group, a separate comparison was performed between varieties for each phase, considering male and female individuals. As a result, DEGs were identified in each phase, separately for male and female individuals, leading to a total of six comparisons within this group. There were no statistically significant DEGs observed in the telogen phase between the female Eskisehir and Lalahan Angora goat varieties. A total of 4 genes (TNNT1, B2M, KRTAP 16.2, and ENSCHIG00000011606) were identified as statistically significantly downregulated with differential expression in the anagen phase between the female Eskisehir and Lalahan Angora goat varieties (Fig. 5).
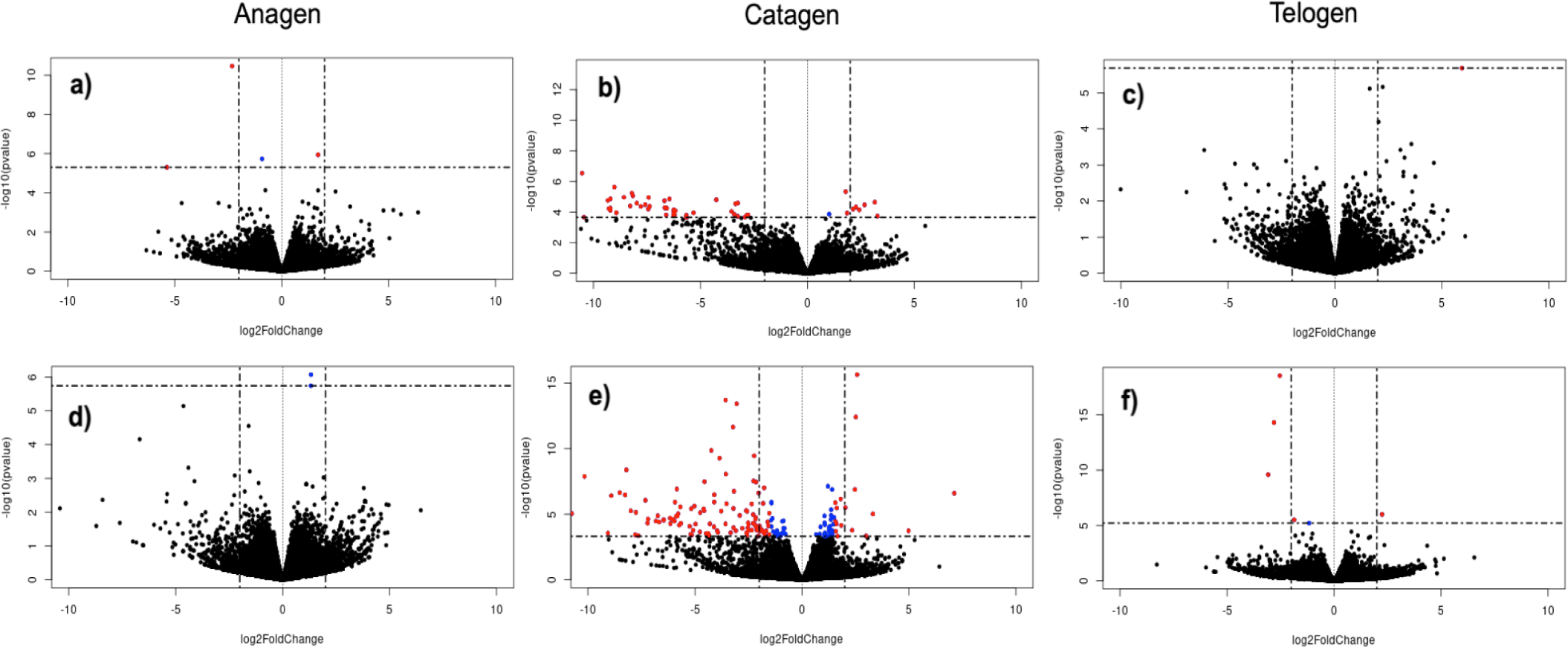
In the catagen phase, 63 DEGs were found between the female Eskisehir and Lalahan Angora goat varieties, with 7 upregulated (LOC108635997: Keratin, type II cytoskeletal 6A-like; LOC102174594: Keratin-associated protein 9-9-like; Keratin-associated protein 4-11; Keratin-associated protein; LOC102171368: Keratin-associated protein 4-9-like; ARHGEF19; and LOC108633884. Supplementary File 4) and 56 downregulated genes (Fig. 6A). On the other hand, there were no statistically significant DEGs observed in the anagen phase between the male Eskisehir and Lalahan Angora goat varieties. A total of five DEGs (TRPM1, KLHL4, ENSCHIG00000011606, ENSCHIG00000018556, and HAL) were found between the male Eskisehir and Lalahan Angora goat varieties in the telogen phase. In the catagen phase, a total of 177 DE genes were found between the male Eskisehir and Lalahan Angora goat varieties. Of these, 47 were upregulated, and 130 were downregulated (Fig. 6B).
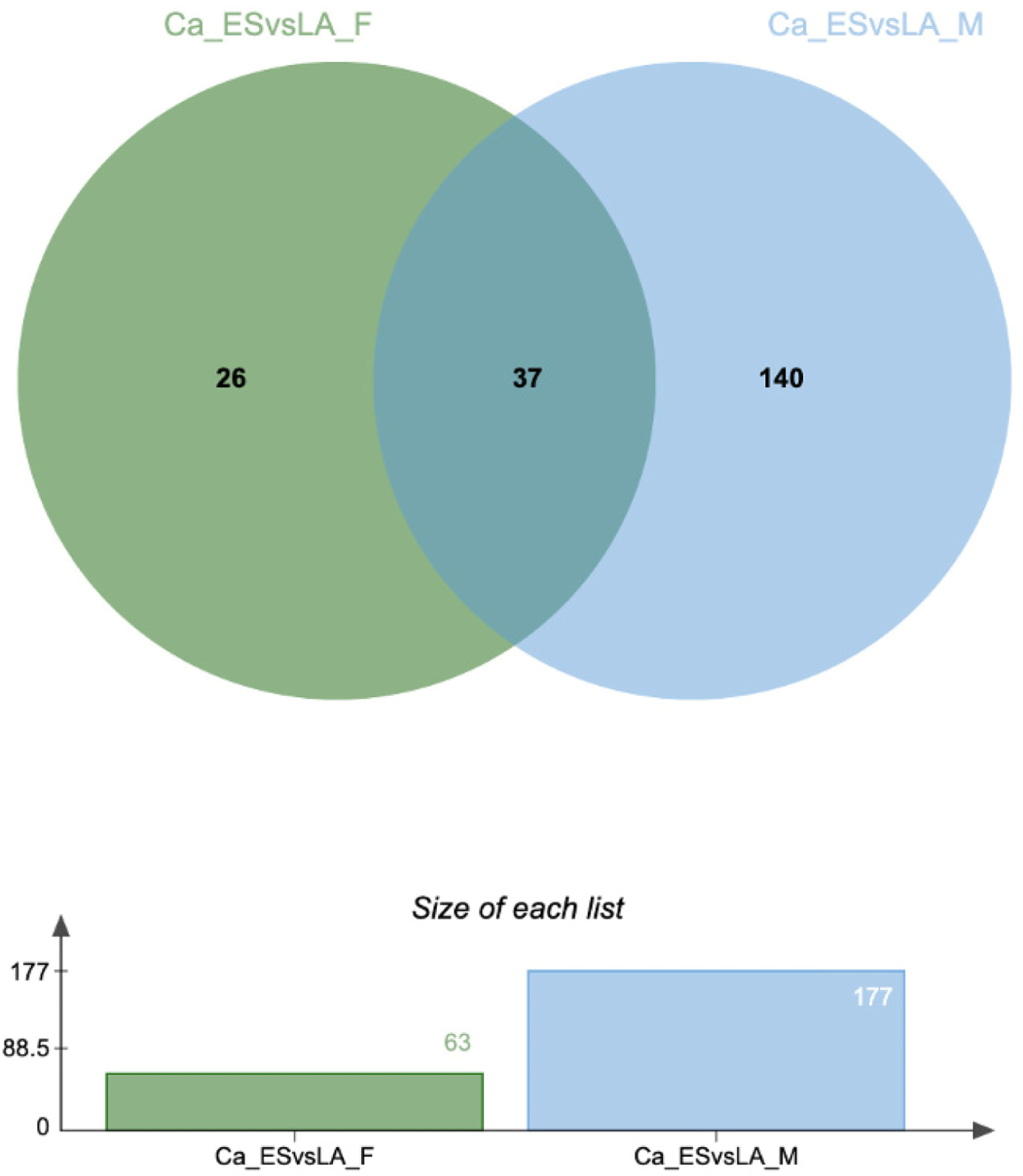
To identify DEGs specific to each goat variety in each phase of hair growth, we compared the DE genes for each variety and sex in each phase. Fig. 7 shows the Venn diagram of the DEGs identified in this comparison group. These findings indicate that the DEGs identified are phase and variety-specific, demonstrating sex-specific patterns as well.
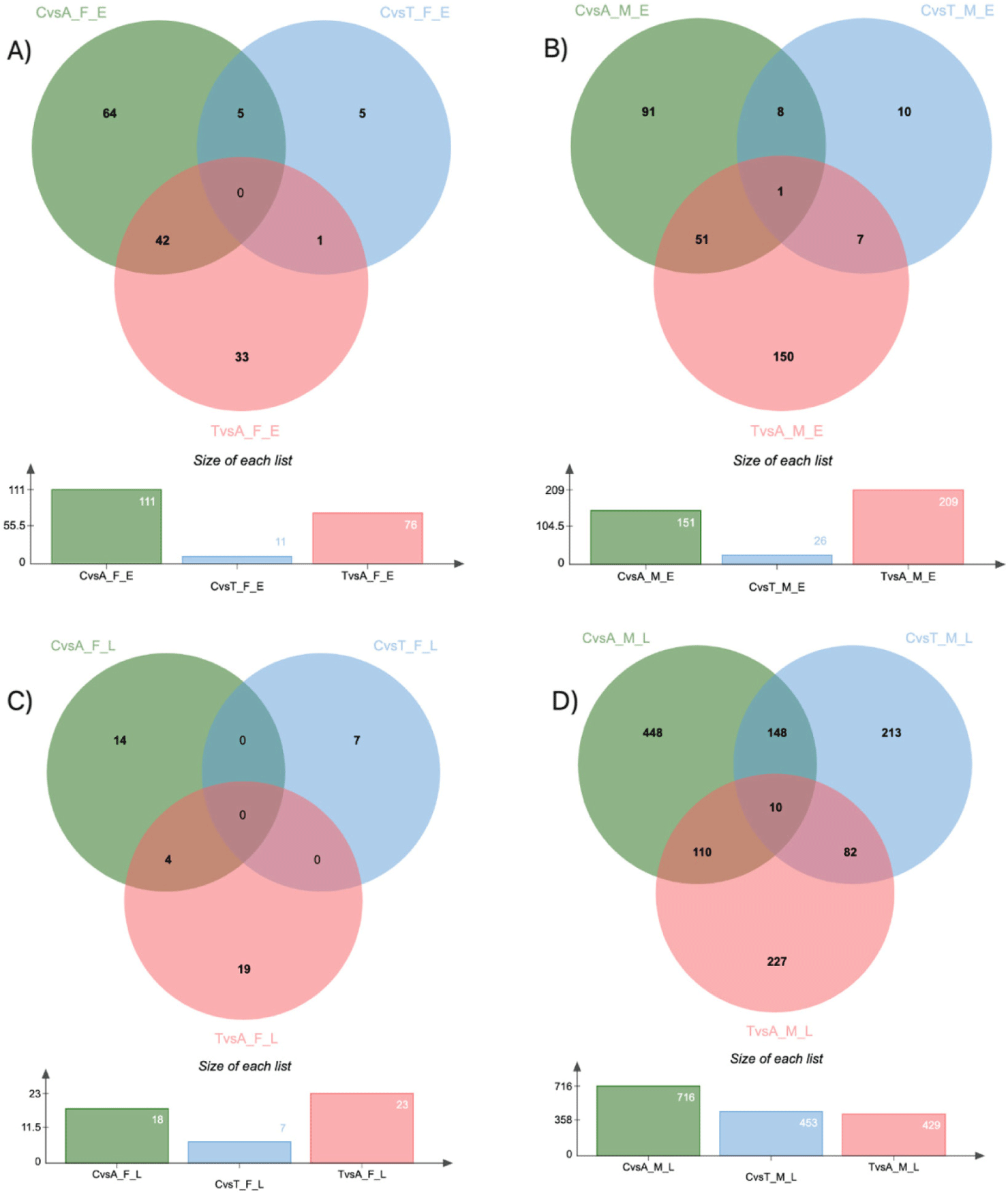
The findings revealed that among the female Eskisehir Angora goat varieties, the DE genes exhibited the greatest variation between the anagen and catagen phases, with a comparatively smaller differentiation observed between the catagen and telogen phases. On the other hand, in the female Lalahan Angora goat varieties, the DEGs in all groups were smaller than those in the Eskisehir Angora goat variety. Our findings suggest that there are significant physiological differences between Eskisehir and Lalahan Angora goats at different stages of hair follicle cycling.
In order to gain a comprehensive understanding of the BPs and pathways related to the development of mohair HF, we carried out GO and KEGG analyses. The top enriched GO terms (p-value < 0.05) were identified by comparing DE across groups. We conducted GO and KEGG pathway analyses on each of the 24 groups, and the obtained results are shown in Supplementary Fig. S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, and S15 and File 5.
In the gender group, the comparison of data between the Eskisehir Telogen Female and Male groups revealed that DEGs were significantly enriched in the keratin filament (GO: 0045095) and intermediate filament (GO:0005882) categories, according to the GO CC analysis. ENSCHIG0000000025280 (LOC106503217: keratin-associated protein 4-9-like), ENSCHIG0000000020416 (LOC108638288: keratin-associated protein 4), ENSCHIG0000000009215, ENSCHIG00000002703, ENSCHIG00000007577 (LOC108638300: Keratin, high-sulfur matrix protein, IIIA3-like) genes that were upregulated were significantly enriched in the keratin filament and intermediate filament categories (Supplementary Fig. S3 and File 5).
The development and cycling of goat HF have been extensively investigated in previous studies. These investigations have shed light on the involvement of multiple key pathways, including tumor necrosis factor (TNF), fibroblast growth factor (FGF) [5], MAPK signaling, WNT pathway [33], bone morphogenetic protein (BMP) family [34], transforming growth factor (TGF) family [35], Sonic hedgehog (SHH) [36], and NOTCH signal transduction pathways [37].
Consistent with the preceding investigations, our results suggest a notable enrichment of the TNF signaling pathway (chx04668) within the comparison group of Lalahan Catagen Female vs. Male (Supplementary Fig. S5 and File 5). ENSCHIG00000007846 (JUNB: JunB proto-oncogene, AP-1 transcription factor subunit), ENSCHIG00000014217 (LOC102182395: growth-regulated protein homolog gamma), ENSCHIG00000025384 (FOS: Fos proto-oncogene, AP-1 transcription factor subunit), ENSCHIG00000021461 (TNFAIP3: TNF alpha induced protein 3), and ENSCHIG00000011290 (SOCS3: suppressor of cytokine signaling 3) genes were significantly enriched in the TNF signaling pathway.
In the variety group, for the Eskisehir Female Anagen vs. Catagen comparison, DE genes for GO BP were significantly enriched in muscle system processes (GO:0003012). For the CC, DEGs were enriched mainly in myofibrils (GO:0030016) and intermediate filaments (GO:0005882). Then, for MF, most of the DEGs were enriched in actin binding and actin filament binding. The KEGG pathway analysis showed that DEGs in comparison between Eskisehir Female Anagen and Catagen were significantly enriched in the Calcium signaling pathway (chx04020) (Supplementary Fig. S6 and File 5). Moreover, out of the 111 genes that were identified through the comparison of Eskisehir Female Anagen vs. Catagen, 94 were found to be orthologous to Homo sapiens. We conducted a Reactome pathway analysis using these 94 genes. The Reactome pathway analysis revealed that the DEGs identified in this group were significantly enriched in the keratinization (R-HSA-680556; KRTAP11-1, KRTAP13-3, KRTAP10-9, KRTAP10-6, KRTAP3-3, KRTAP4-5, KRT33A, KRTAP3-1) pathway (Fig. 8).
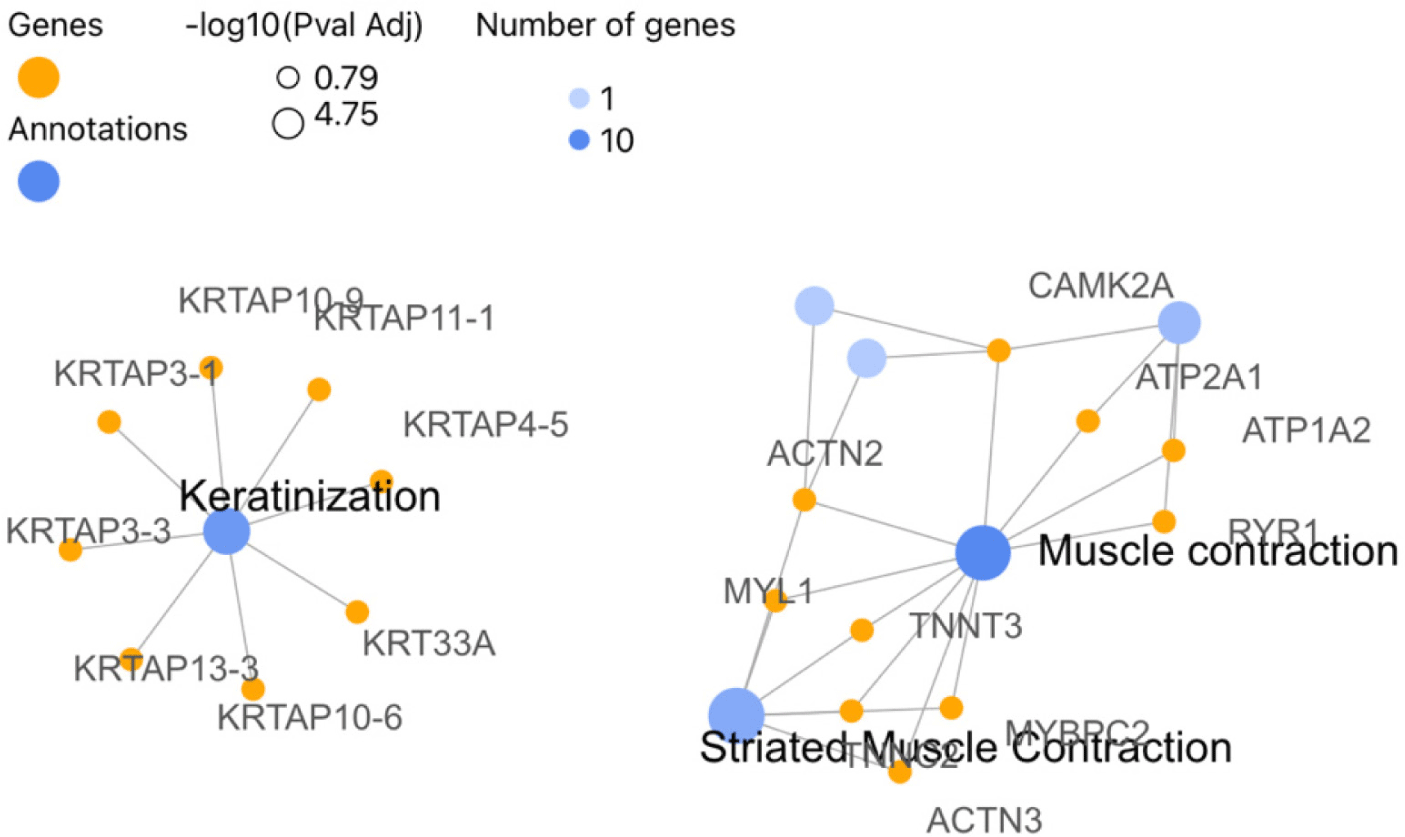
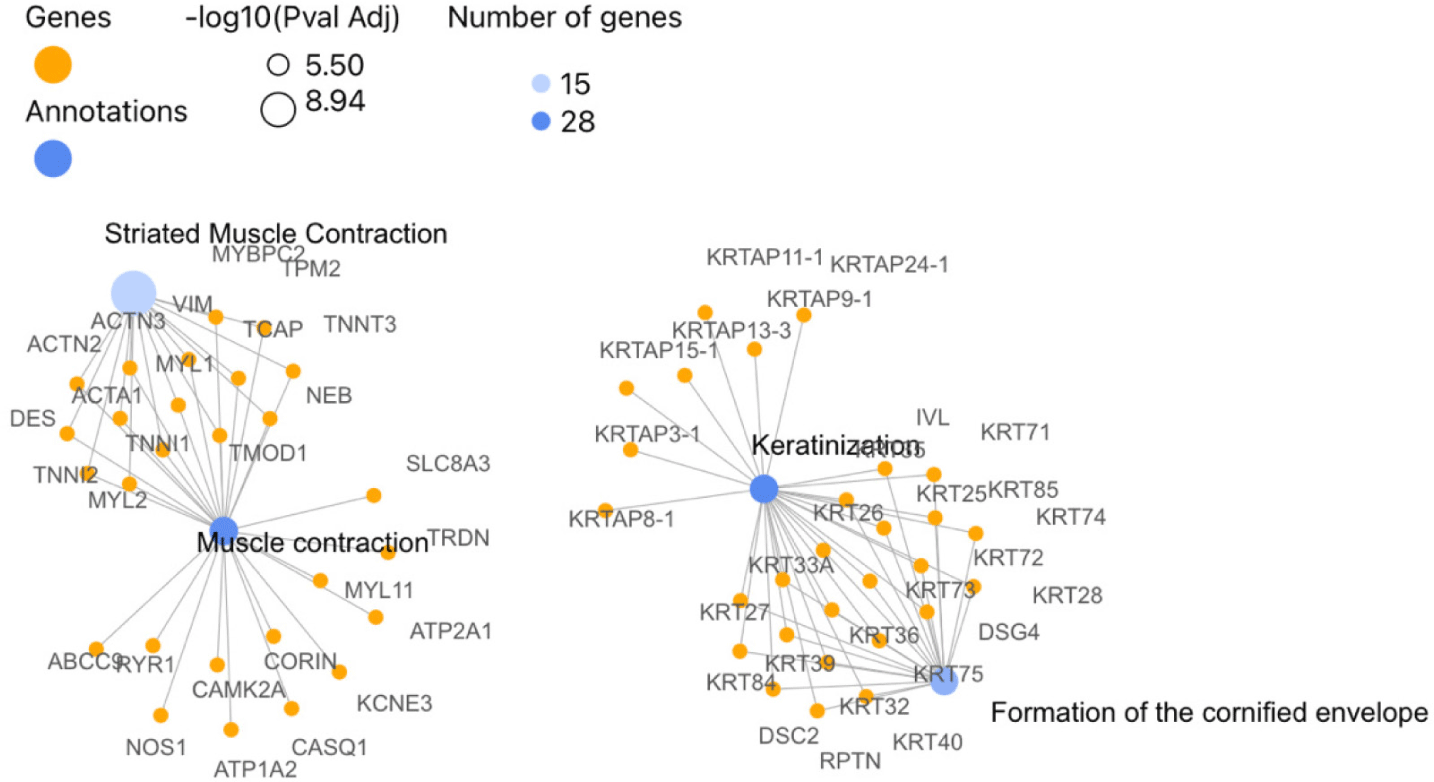
In the Lalahan variety of Angora goat, 716 DEGs were identified between anagen and catagen phases in males, out of which 654 were found to be orthologous with humans. In contrast to the results of the Eskisehir Female Anagen and Catagen comparison, the Reactome pathway analysis revealed that the DEGs in the Lalahan Male Anagen vs. Catagen comparison group were significantly involved in the “Keratinization” (R-HSA-6805567), “Muscle Contraction” (R-HSA-397014; 27 genes), and “Formation of the Cornified Envelope” (R-HSA-6809371; 21 genes) pathways (Fig. 9). On the other hand, DE genes for GO BP were significantly enriched in “muscle structure development” (GO:0061061, 57 genes) and “actin filament-based processes” (GO:0030029). For the CC, DEGs were enriched mainly in “keratin filament” (GO:00045095) (Table 1) and intermediate filament (GO:0005882) (Supplementary Fig. S11 and File 5).
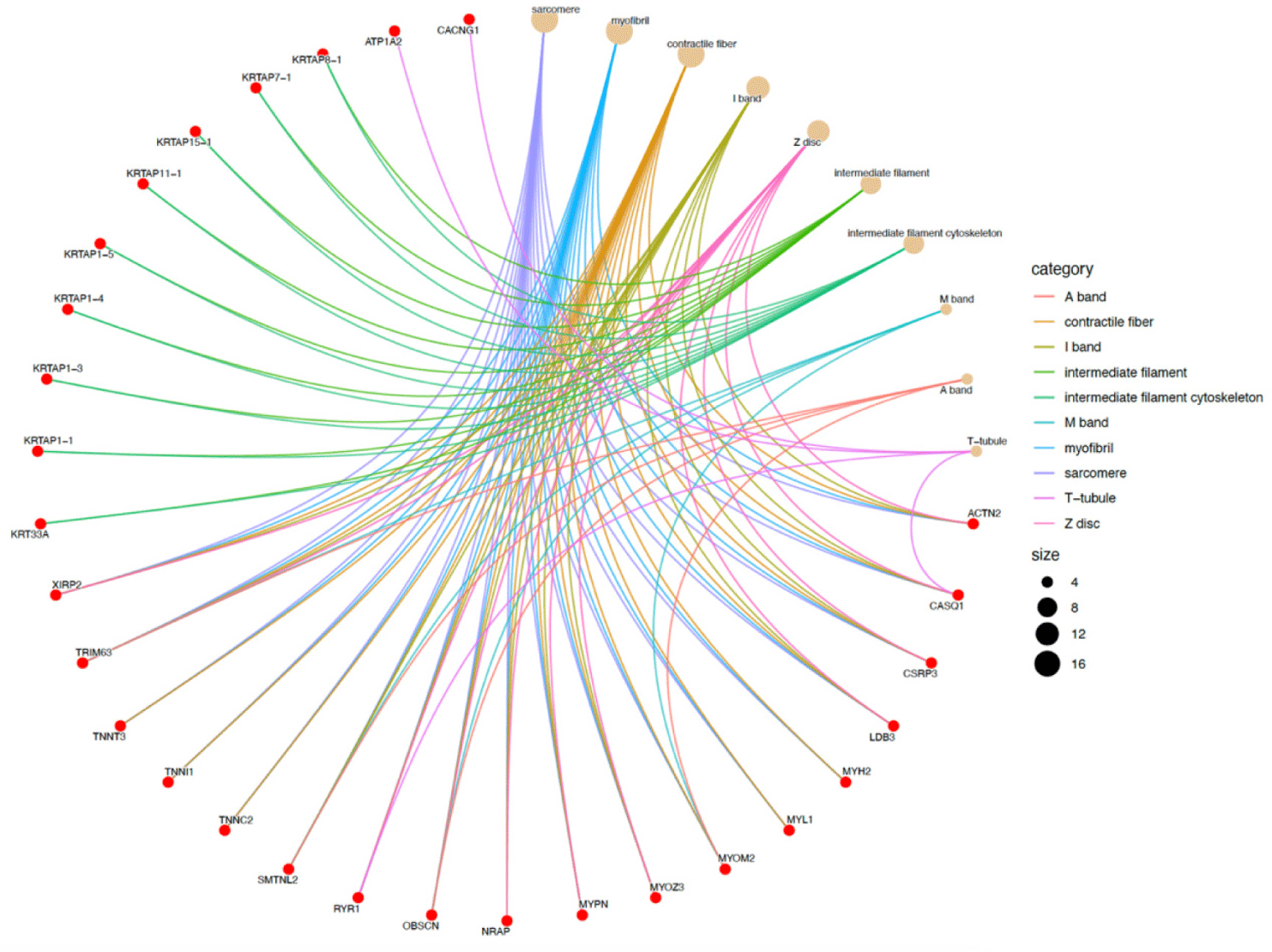
In the Eskisehir Female Anagen vs. Telogen comparison, the DEGs involved in GO BPs were significantly enriched in “actin-myosin filament sliding” (GO:0033275) and “myofibril assembly” (GO:0030239). For the CC, the DEGs were mainly enriched in “myofibril” (GO:0030016; 18 genes) and “intermediate filament” (GO:0005882; KRT33A, KRTAP1-1, KRTAP1-3, KRTAP1-4, KRTAP1-5, KRTAP11-1, KRTAP15-1, KRTAP7-1, KRTAP8-1). A detailed cnetplot visualization of these CC processes is presented in Fig. 10.
The Lalahan Female Anagen vs. Catagen, Anagen vs. Telogen, Catagen vs. Telogen, and Eskisehir Female Catagen vs. Telogen groups were excluded from GO and pathway analysis due to the limited number of genes in these groups.
In the inter-variety group, for the Catagen Female Eskisehir vs Lalahan comparison, DEGs for GO CC were significantly enriched in “myofibril “(GO:0030016, 22 genes) and “keratin filament” (GO:0045095). For MF, most of the DEGs were found to be enriched in the “process of acting binding” (GO:0003779, 14 genes) (Supplementary Fig. S14 and File 5). For the Catagen Male Eskisehir vs. Lalahan comparison, DE genes for GO BP were significantly enriched in “muscle structure development” (GO:0061061, 18 genes), For the CC, DEGs were significantly enriched in the “keratin filament” (GO:0045095, 19 genes). The KEGG pathway analysis showed that DEGs in the comparison between Catagen Male Eskisehir and Lalahan were significantly enriched in the Metabolic pathway (chx01100) (Supplementary Fig. S15 and File 5). Moreover, to identify DEGs specific to each gender in the catagen phase of hair growth for each goat variety, we compared the DEGs for each variety and sex in this phase (Fig. 11). The results showed that KRT40, KRT72, KRTAP10, KRTAP9, KRTAP2, KRTAP3, and KRTAP8 were only DE in the Male catagen Eskisehir vs. Lalahan group, while KRT6 was only DE in the Female catagen Eskisehir vs. Lalahan group. This information can be used in breeding selection.
To confirm the results of the transcriptomic analysis, we randomly selected five genes with differential expression and performed a qPCR analysis. The genes chosen for analysis —FGF5, FOS, KRTAP8, FOSL1, and FOSB—exhibited a consistent expression pattern in both the qPCR results and RNA-Seq data, thereby confirming the reliability of our transcriptome information (Supplementary Fig. S16).
DISCUSSION
Within a dynamic environment, the hair follicle, a functional miniature organ, undergoes development that is influenced by various molecular signals. The hair cycle, which consists of three distinct phases, controls the growth, regression, and resting periods of HF. This cycle serves as an important model for investigating the regulation, activation, and quiescence of stem cells, as well as the processes of cell proliferation, differentiation, and apoptosis within adult regenerative epithelial tissue.
Insufficient knowledge exists regarding the alterations in gene expression that occur during the transition from anagen to catagen and telogen in the Angora goat and its specific varieties. To advance our understanding of the molecular mechanisms underlying this transformation, it would be beneficial to conduct global gene expression measurements and perform differential gene expression profiling of distinct HF. As of our current knowledge, the application of RNA-Seq technology for analyzing HF in Angora goats and their specific varieties has not been reported to this date.
In this study, according to the comparison results of males and females in each phase and variety, a higher number of DEGs were found in the anagen phase than in the catagen and telogen phases, while the lowest number of DEGs were found in the telogen phase. In contrast to our results, Su et al. observed distinctive gene expression patterns in Inner Mongolian Cashmere goat skin samples across various stages of the hair growth cycle [38]. Notably, they identified 51 DEGs between the anagen and catagen stages, 443 DEGs between the catagen and telogen stages, and 779 DEGs between the telogen and anagen stages. These findings provide insights into the molecular mechanisms that lie under the hair growth cycle in this specific breed of goats. The authors emphasized that the most substantial number of DEGs was observed between telogen and anagen, whereas the smallest number of DEGs was identified between anagen and catagen. The contradictory results of these studies confirm that hair follicle development in goats may vary according to breeds. However, our results are consistent with the process of the hair growth cycle. A HS is produced by the follicle from tip to root during the growth period, or anagen. The hair follicle experiences epithelial proliferation and differentiation during the anagen phase. During this stage, the IRS undergoes keratinization, a process that imparts structural stability and direction to the growing HS. In contrast, the catagen phase represents a transitional stage where the follicle prepares for the telogen phase, which is a period of dormancy where no new hair is produced. The transition from the anagen phase to the catagen phase is of utmost importance in the hair cycle. In this phase, the matrix cells, which are responsible for hair growth, undergo a restricted number of cell divisions before initiating the process of differentiation. As the availability of matrix cells decreases, the process of differentiating the HS and IRS slows down, leading the follicle into a destructive phase known as catagen. Subsequently, the HF enter a resting phase called telogen. The activation of one or two quiescent stem cells, located at the base of the telogen follicle near the DP, is accountable for the transition from telogen to anagen. This transition leads to the formation of a new HS. The cells initiate a rapid proliferation process, resulting in the rapid multiplication of transit-amplifying daughter cells that will eventually give rise to the formation of the new hair follicle. This newly formed follicle takes shape adjacent to the existing pocket that houses the club hair, which will eventually be shed. As a consequence, a bulge is formed, comprising a fresh layer of stem cells that play a crucial role in replenishing the stem cell reservoir. Interestingly, the emerging hair emerges from the same upper opening as the previous hair. The transition from the telogen to anagen phase bears similarity to the activation of embryonic skin stem cells, wherein they are stimulated to generate a new hair follicle from scratch. Most of signaling molecules that regulate hair follicle morphogenesis belong to Wnt pathway, FGF family, TNF family, BMP family, SHH transduction pathway, TGF family and Notch transduction pathway. Some of them show activator and some inhibitory effects. However, the exact nature, timing, and intersections of these signaling pathways remain unclear. [39].
The expression levels of most genes in the skin were correlated with the activity of the hair follicle, suggesting that the two are closely linked. Therefore, these results suggest that there were probably gradual changes in the skin between male and female Angora goats as the hair follicle moved from the growth phase to the resting phase.
Similarly, in the variety group, the gene expression patterns in the male groups appeared to be significantly different from those in the female groups when comparing each hair growth phase. This result confirms the effect of androgens and estrogens on the hair growth process. It’s been known for over 70 years that estrogens play a role in skin physiology and the control of hair growth. Previous research has established a connection between sex steroid hormones and the regulation of different aspects of skin structure and function. These hormones play a role in controlling processes such as hair growth, pigmentation, vascularity, water retention, and elasticity. The current understanding of sexual hair growth acknowledges the involvement of both adrenal androgens and ovarian hormones in human females. It is no longer limited to just adrenal androgens. Estrogens have the ability to directly stimulate hair growth at the hair follicle level in the skin, either independently or in conjunction with androgens. Furthermore, estrogens serve as effective regulators of hair follicle growth and cycling [40,41]. Androgens are responsible for anagen shortening or premature catagen entry [41]. The relationship between sex and hair growth in goats, as well as the underlying molecular mechanisms, are worthy of further investigation.
Moreover, Fig. 7 visually depicts the complexity of DEGs identified in this study through a Venn diagram. These findings highlight the phase and variety-specific nature of DEGs, with an additional layer of sex-specific patterns. Notably, female Eskisehir Angora goats displayed the most variation in DEGs between the anagen and catagen phases, suggesting a crucial transition in gene expression during this active hair growth stage. Interestingly, the differentiation between catagen and telogen phases was less pronounced. Conversely, female Lalahan Angora goats exhibited a consistently lower number of DEGs across all phases compared to Eskisehir goats. This observation suggests significant physiological differences between these two varieties, potentially influencing hair follicle cycling at the molecular level. Further investigation into the specific genes and pathways involved in these breed-specific variations could provide valuable insights into the regulation of hair growth in Angora goats. Further delving into sex-specific differences, Fig. 11 explores DEGs specific to the catagen phase for each goat variety. Interestingly, the analysis revealed a cluster of keratin-associated protein genes (KRT40, KRT72, and KRTAP10, etc.) solely DE in the male Eskisehir versus Lalahan comparison. This suggests potential sex-specific regulatory mechanisms involving keratinocyte differentiation during the catagen phase, which might be unique to the Eskisehir variety. Conversely, only KRT6 emerged as a DE gene specific to the female Eskisehir versus Lalahan comparison. These findings highlight the intricate interplay between sex, breed, and specific hair growth phases. Additionally, the identification of these DEGs holds significant value for breeding programs. By incorporating this information into selection strategies, breeders can potentially target specific traits related to hair quality and growth patterns in Angora goats.
In the female, when we compared the catagen and telogen phases, we observed that the expression of genes for keratins and KAPs (KRTAP8-1, KRTAP16.4, and KRTAP21-1) was quite higher in Eskisehir Angora goats. Interestingly, KRT21-1 was found to be expressed as four different protein isoforms: ENSCHIG00000010356, ENSCHIG00000009372, ENSCHIG00000015280, and ENSCHIG00000011204. The UniProt accession numbers for these genes are A0A452E3E7 (93 aa), A0A452E013 (90 aa), A0A452EQT5 (87 aa), and A0A452E733 (87 aa), respectively. This may indicate that alternative isoforms of KRT21-1 may be functional in the transition from catagen to telogen. Further studies are necessary to confirm the roles of the KRT21 gene in hair follicle development.
As the hair cycle transitions from anagen to catagen, apoptosis of epithelial cells in the hair strand begins. These apoptotic cells are then phagocytosed by macrophages and neighboring epithelial cells. Throughout this process, keratin remains the major protein in the hair. Consistent with this information, for the Eskisehir Female Anagen vs. Catagen comparison, we found that DEGs identified in this group were significantly enriched in the keratinization pathway (R-HSA-680556; KRTAP11-1, KRTAP13-3, KRTAP10-9, KRTAP10-6, KRTAP3-3, KRTAP4-5, KRT33A, KRTAP3-1) for Reactome. We also found that DEGs in this group were significantly enriched in the “Calcium signaling pathway” for KEGG analysis. A total of 7 DEGs (ATP2A1, TNNC2, FGF21, HRC, CAMK2A, RYR1, CACNA1S) were involved in the calcium signaling pathway chx04020 (Supplementary Fig. S17), which may have a potential indirect role in MAPK signaling and apoptosis pathways. The MAPK signaling pathway and the cell cycle pathway, among others, also play an important role in hair follicle development [20]. The enrichment of DEGs in the MAPK signal pathway suggests that these genes may have a role in cell proliferation and differentiation, which are essential processes in hair follicle growth and development. This is consistent with previous findings that the MAPK signal pathway is an important regulator of hair follicle growth and development in goats [38,42]. In the anagen-catagen comparison, in contrast to Eskisehir variety, we found that DEGs identified in Lalahan male group were significantly enriched in the keratinization pathway (R-HSA-6805567; KRTAP15-1, KRTAP11-1, KRT75, KRT40, KRT28, KRT32, DSG4, KRT36, KRTAP13-3, KRT73, KRT72, KRT26, KRT85, KRT39, KRTAP24-1, KRTAP8-1, KRT33A, KRT71, RPTN, KRT74, KRTAP3-1, DSC2, KRT25, KRT84, KRTAP9-1, IVL, KRT27, KRT35) for Reactome and all these genes were found as downregulated. The downregulation of KRT and KRTPs suggests a significant decrease in keratinization from the anagen to catagen transition, consistent with the molecular process of hair growth. The transition from the anagen to the catagen of the hair follicle may indicate that gradual changes are taking place within the skin. Additionally, the internal molecular microstate of the skin undergoes a significant change when the hair follicle enters a new cycle of growth from the resting phase. Furthermore, the findings of this study are consistent with those of [43], who suggested that KRT84 and KRT25 can be considered as candidate genes for hair follicle morphogenesis. Additionally, according to the GO CC results, 30 DEGs were enriched mainly in keratin filament. As a result, we determined that KRT and KRTAPs were highly functional and had quite different expression patterns between female and male goats, as well as between Eskisehir and Lalahan Angora goats.
The results of comparing varieties for each phase, considering male and female individuals, show that DEGs were only determined in the catagen phase in both female and male goats. For the Catagen Female Eskisehir vs. Lalahan comparison, DEGs for GO CC were significantly enriched in “keratin filament” (GO:0045095, ENSCHIG00000025429: LOC108635997 keratin, type II cytoskeletal 6A-like, ENSCHIG00000020255: LOC102174594 keratin-associated protein 9-9-like, ENSCHIG00000023156, ENSCHIG00000015597, ENSCHIG00000017056: LOC102171368 keratin-associated protein 4-9-like).For the Catagen Male Eskisehir vs Lalahan comparison, DE genes for GO CC, DEGs were enriched significantly in keratin filament (GO:0045095, 19 genes: ENSCHIG00000010601, ENSCHIG00000006780, ENSCHIG00000021086, ENSCHIG00000006463:LOC108638298: keratin, high-sulfur matrix protein, IIIA3-like; ENSCHIG00000000533:LOC108636561: keratin-associated protein 10-11-like; ENSCHIG00000010344:LOC108636554: keratin-associated protein 10-8-like; ENSCHIG00000023156, ENSCHIG00000007612:LOC108636430: keratin-associated protein 10-12-like; ENSCHIG00000008117:LOC108636431: keratin-associated protein 12-2-like; ENSCHIG00000001659:KRTAP3-1; ENSCHIG00000011548:LOC108636548: keratin-associated protein 10-1; ENSCHIG00000004564:LOC102170546: keratin-associated protein 3-3; ENSCHIG00000015017:LOC108636550: keratin-associated protein 10-8-like; ENSCHIG00000024813:LOC102172766: keratin-associated protein 10-11-like; ENSCHIG00000026079, ENSCHIG00000011563:LOC100861174: keratin associated protein 12.1; ENSCHIG00000026525:LOC102177517: keratin, type II cytoskeletal 72; ENSCHIG00000026301:LOC102184693: keratin, type II cuticular Hb5; ENSCHIG00000026924:LOC108638297: keratin, high-sulfur matrix protein, B2C). The KEGG pathway analysis showed that DEGs in the comparison between catagen male Eskisehir and Lalahan goats were significantly enriched in the metabolic pathway, while they were significantly enriched in the calcium signaling pathway in female goats. During the catagen phase, hair growth ceases and several morphological changes occur, such as DP condensation and movement upwards. Additionally, cytological changes may also occur, including apoptosis of epithelial cells and the ORS. Catagen, the intermediate phase between anagen and telogen, plays a crucial role in the hair growth cycle. During this phase, the lower portion of the hair follicle regresses entirely, including the bulb and ORS. This intricate process is characterized by apoptosis, or programmed cell death, of the epithelial cells within these layers [39,44,45]. The findings presented in this study indicate that distinct molecular pathways governing regulation are implicated in the hair follicle cycle of Eskisehir and Lalahan Angora goats. These pathways exhibit variations not only among different varieties but also between males and females.
Fibroblast Growth Factor 5 (FGF5) belongs to the FGF gene family and is primarily expressed during the anagen phase of the hair cycle. Its main function is to regulate the transition from the growth phase (anagen) to the resting phase (telogen) [39,46]. Research has indicated that the absence of the FGF5 gene leads to a prolonged anagen phase during hair growth. In a study conducted by Su et al. it was observed that FGF5 and FGFR1 had an impact on the growth cycle of HF in Inner Mongolian Cashmere goats [38]. Furthermore, Zhang et al. utilized RNA-seq to analyze genome-wide expression and investigate DEGs associated with the cycling of HF in cashmere and milk goat breeds across various seasons [42]. The researchers discovered that the FGF5 gene exhibited its lowest expression levels during December, which corresponds to the catagen phase, while its highest expression levels were observed in September, during the anagen phase, in milk goats. This finding supports the notion that the transition from anagen to catagen occurs between the fall and winter seasons. Additionally, the expression patterns of FGF2, FGF8, and FGF22 mirrored those of FGF5, with their highest expression levels occurring in September (anagen) and lowest in December (catagen). Furthermore, a significant number of FGF genes demonstrated their highest expression levels in March (telogen), while some genes exhibited high expression levels in both March and June. The researchers claim that the discrepancy in the quantity of DEGs observed in September and December in cashmere goats, in contrast to the higher number of DEGs found in milk goats, provides compelling evidence that cashmere goats do not experience the anagen to catagen transition during the transition from autumn to winter. The evidence for this conclusion is reinforced by the increased level of FGF5 expression observed in December, which closely resembles that of September. Additionally, the expression of telogen FGF18 was significantly elevated in September, further supporting the aforementioned conclusion.
Moreover, the distinctively high levels of FGF8 and FGF22 expression were observed exclusively in the identical group of September milk goats, and this elevated expression pattern persisted in both September and December cashmere goats. Moreover, TP53, an inhibitor of anagen, demonstrated the most significant level of expression during the month of September. Additionally, the authors highlighted that the expression profiles of FGF5 in milk goats suggest that the HF undergo both catagen and telogen phases during winter, while anagen begins in March and concludes in autumn [42]. Our findings indicate that the FGF5 gene exhibited a significant differential expression exclusively in male Lalahan Angora goats during the transition from telogen to anagen. This observation aligns with the notion that the FGF5 gene plays a crucial role in this specific stage of hair growth in male Lalahan Angora goats.
CONCLUSION
Mohair fiber is a valuable animal fiber source. Despite the significance of mohair, existing literature has predominantly focused on cashmere fiber, with limited studies dedicated to mohair. This study is the first to evaluate the impact of genes on hair follicle development in Lalahan and Eskisehir variety Angora goats using RNA sequencing methods. Consistent with the hypothesis, the study revealed notable differences between the two varieties in terms of hair follicle development. Specific genes were identified that play distinct roles in the hair growth cycles of Lalahan and Eskisehir Angora goats, contributing to our understanding of the molecular mechanisms underlying mohair production. This study has certain limitations, including the sample size and the scope of gene expression analysis. Future studies with larger sample sizes and broader genomic analyses are necessary to validate and expand upon these findings. The discoveries made in this study highlight the genetic differences influencing hair follicle development in Angora goats, which can inform breeding programs aimed at improving mohair quality. Understanding these genetic factors can lead to enhanced selection strategies for desirable fiber traits. Further investigations are needed to explore the functional roles of the identified genes and their interactions in hair follicle development. Additionally, studies examining environmental and management factors in conjunction with genetic data will provide a more comprehensive understanding of mohair production. This study is expected to serve as a valuable resource for future research endeavors in the field of animal fiber genetics, paving the way for advancements in mohair quality and production.
