Background
Methane (CH4), one of the three main greenhouse gases (GHG) besides of carbon dioxide (CO2) and nitrous oxide (N2O), have a global warming potential of 28-fold than that of carbon dioxide (CO2) [1]. Agricultural sector is considered to contribute the biggest methane emission, which calculated around 50.6% from anthropogenic methane [2]. Within agriculture, the livestock sector contributes approximately 18% of the global anthropogenic GHG emission [3]. Among livestock, ruminant contributes about 81% of GHG [4] due to massive methanogenesis by rumen microbes, which produce 90% of total CH4 production from ruminants [5]. Globally, CH4 emissions of dairy and beef cattle denote 30% and 35% of the livestock sectors’ emissions. However, buffalos and small ruminants are lower contributors, demonstrating 8.7% and 6.7% of sector emissions, respectively [6]. The CH4 production in ruminants represents a gross energy loss from 2% to 14% of gross energy consumption [7]. Therefore, reduction of methane emission in animal conserves an energy and enhances productivity.
For the fulfilling of nutrition demand of growing population, the number of domesticated animals increasing rapidly. This high number of animals is directly proportional to CH4 production. In developed countries, it often recommends culling of nonproductive and low-producing animals to reduce CH4 budget [8]. They maintained high-producing animals in herds for reducing CH4 emissions per unit of product. Conversely, this is often impractical for poor countries due to their socioeconomic and religious background. It is well established that with the increasing animal productivity, CH4 emissions also decrease per unit of products. There are many options for enhancing the productivity of animals such as the proper formulation of diets, supplementation of protein and energy to low-quality forages, ionophores, bovine somatotrophin, and probiotics [9]. Lately, both the increasing of animal production as well as decreasing the methane emission by the animal especially ruminants are the main focus among researchers throughout the world. A number of CH4 abetment strategies from ruminants have been revised earlier [2,8,10–26]. However, these strategies summarized more concreate and cumulative approaches to set up future research needs for sustainable methane mitigation strategies in ruminants focusing direct-feed microbials. As a part of cumulative approach, accurate estimation of methane production is also very important in order to make a suitable methane mitigation strategy. Therefore, this review also summarized the methods of enteric methane measurement and their applications.
The rumen microbiome including a wide variety of microorganisms, viz. bacteria, archaea, ciliated protozoa, fungi, and viruses, stay in a symbiotic relationship in a strict anaerobic condition within the rumen [27]. The protozoa can comprise up to 50% of the microbial biomass in rumen [28]. While, the fungi were estimated at around 8% of the total biomass [29] but may reach 20% in sheep [30]. The archaea include only 0.3%–4% [31], and the bacteria cover the remainder, characteristically the largest component of the rumen microbial biomass [26]. This rumen microbiome plays a significant role in feed fermentation within the rumen and produces different volatile fatty acids (VFAs), CO2 and H2. These VFAs are essential for energy metabolism and protein synthesis of the ruminant host [32]. Among the diverse rumen microbiomes, only a few of these have been successfully characterized earlier based on culture-techniques. Recently, the application of multi-omics techniques such as metagenomics by next-generation sequencing (NGS) or high-throughput sequencing [33–37], metatranscriptomics [38–40], metaproteomics [41,42], and metabolomics [43–45] have been increased greatly [40].
Methanogenesis is a process of CH4 production in the rumen where H2 reduced the CO2 with the help of methanogenic archaea [46]. CH4 production is the main way for H2 clearance from fermentation [47]. There are two main pathways of methanogenesis in the rumen, carried out by archaea, are presented in Table 1. The hydrogenotrophic pathway converts H2 and CO2 into CH4 by the bacteria, protozoa, and fungi [5,23]. It is usually implicit that formate can be used by most abundant ruminal archaea that equivalent to H2+CO2, so formate is included in the hydrogenotrophic category [31,48]. Methylotrophic pathway is an another pathway of methanogenesis, which use methyl groups such as those present in methylamines and methanol as substrate [26,49,50]. The methanogens species have classified into 28 genera and 113 species, but it can be expected to occur many more in nature [15,31]. From rumen, only a few methanogens have been so far isolated based on culture techniques such as Methanobrevibacter ruminantium, Methanobrevibacter millerae, Methanobrevibacter olleyae, Methanobacterium formicicum, Methanobacterium bryantii, Methnaomicrobium mobile, Methanoculleus olentangyi, Methanobrevibacter smithii and Methanosarcina spp. [31]. Lately, multi-omics techniques are using to understand greenhouse gas emission from ruminant production [51].
Accurate estimation of enteric methane production from animals is the key to take initiative for the setting up of mitigation strategies. Mitigation strategy often was unsuccessful due to wrong measurement of CH4 production. A number of enteric methane measuring techniques have been developed. However, respiration chamber (RC), sulfur hexafluoride (SF6) tracer technique, and the automated head-chamber system (AHCS) (GreenFeed; C-Lock Inc., Rapid City, South Dakota, USA) were used successfully and widely in various experiments focusing dairy or beef cattle in several environmental conditions [18,52]. All three methods have been effectively used in a large number of trials with dairy or beef cattle under a wide variety of environmental conditions. However, inconsistent results also observed while did comparative study among techniques [53]. The several enteric CH4 measurement approaches are presented in Fig. 1.
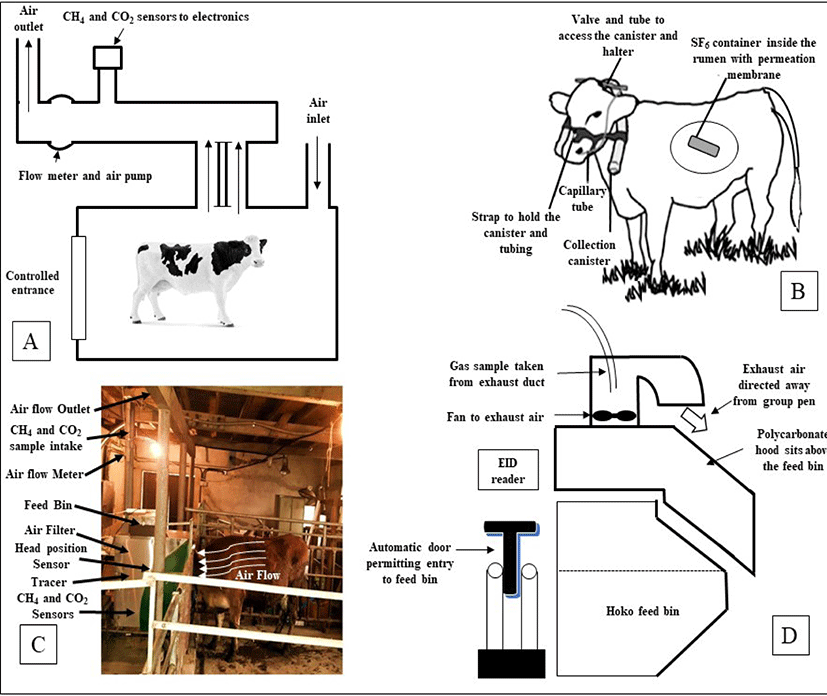
Exact measurements of CH4 emission can be gained by housing animals in RC, which then allow measurement of total methane emission directly. Reynolds et al. [54] and Cammell et al. [55] described the details of RC and measurements of methane emission. For the measurements of gaseous emissions, two open-circuit RC was used (internal volume approximately 21 m3), with airlocks permitting access for faecal and urine collection [55]. An integrative sample of ambient and RC exhaust air was analyzed at 4-min intervals, and there was a switch to calibration gases (oxygen-free nitrogen and nitrogen carrier with 20.5%, 3000 ppm, and 200 ppm oxygen, carbon dioxide, and methane, respectively) every 4 h to provide gas analyses with variation coefficients of 5% or less. This technique is relatively expensive, and are troublesome for the animal to behave normally like a natural that occurs within grassland environments.
The SF6 tracer technique [56,57] can be used to make estimations of methane emissions either by eructation or expiration from animals that can easily select their diet in a way characteristic of farmed livestock especially in grazing. The SF6, a gas is easily measurable and traceable at low concentrations. In addition, it is synthetic in origin and not produced as part of any sort of biological process. The SF6 is also idyllic as its background concentration, which is naturally very low (6 pmol/day) [58], while its concentration as a tracer typically ranges from 0.01 to 0.03 mmol/day [59]. In the gas technique, SF6 tracer gas is delivered via a permeation tube, which is positioned in the rumen, and the ratio of CH4 to SF6 in the breath of an animal is measured and corrected with reference to the background concentration. Though the concentration of the tracer is known, the rate of production of CH4 can be calculated [47]. The assessments have challenged the accuracy of the SF6 technique for estimating CH4 emissions [60,61], with greater between-animal variation compared to RC [62]. The SF6 technique has also delivered variable estimates of CH4 emission from animals on diverse herbages that have not been supported by RC measurements [63–66]. There was also some problem with halter and collection canisters, placed on the animal for CH4 estimates can affect during grazing [67], especially in young animals, and a lower than predicted feed dry matter intake (DMI) will overestimate CH4 yields (g/kg DMI). Administration of rumen SF6 boluses and frequent handling of animal is needed which can be troublemaking to normal behavior and is relatively laborious. Though this technique is challenging to use in practice, standard operating techniques have been established [68].
In 2010, C-Lock Inc., Rapid City, South Dakota, USA has introduced the commercial GreenFeed (GF) system as a static short-term measurement device that measures CH4 emission from distinct cattle and uses head position sensors in combination with decision rules to assess the validity of measures obtained [52,69]. Depending on the experimental design, this GF system can be installed both conditions either grazing field or inside the farm. The animal remaining free to move about and voluntarily enters into the hood where a feed supplement is dropped. The measurements of CH4 emission by the GF system can be done typically over a short period (3–7 minutes), several times within a day, over several days. The GF system estimated CH4 emission using sensors that identified the animal and its head position within a sampling hood, air flow, and CH4 and CO2 concentrations in exhaust air. A radio frequency identification (RFID) reader recognized the animal’s ear tag and GF sampling was activated when the animal’s head was in the correct position within the hood. Satisfactory animal head position resulted in the dispensing of feed pellets which influenced the animal to maintain a suitable head position for accurate measurements. Only one cattle can visit one GF unit at a time point and a ‘visit’ is defined as a visit that results in a methane measurement. The system is automatically programmed using C-Lock Inc. software to control the timing of feed availability for each animal and thus, encourage animals to allocate their voluntary GF visitation, and measure CH4 emission over a 24 h period. Each GF unit can be used for several animals, with manufacturer recommendations of 15–20 animals/unit when grazing and 20–25 animals/unit if housed in free stalls. Cattle are voluntarily participating to visit the GF unit if they adapt once.
The face-masks method is one of the oldest technique for “spot-sampling” of respiratory exchange and CH4 emission from cattle, sheep, and goats. Face mask is only useful for short-term measurements of CH4 emission rate for screening of large numbers of animals, however may cause marked discomfort and distress which can change animal behaviors, and affect the gas measurements [70]. The sniffer method, first reported by Garnsworthy et al. [71], is the measurement of CH4 concentration in air eructed by cattle during milking. In this technique, air in the manger is continuously sampled, analyzed, and logged at 1-second intervals using data loggers in order to measure CH4 and CO2 concentrations in close proximity to the muzzle of the animal. Garnsworthy et al. [71] also reported a good relationship (r = 0.79) between RC technique and CH4 emission rate using this method. The hand laser CH4 detector (LMD) has been proposed to measures enteric CH4 concentrations in the air near the nose or mouth of an animal in normal environment [72]. The methane hood (MH) system, a novel method to quantify CH4 emission from cattle during group feeding in housed environment. This system measures CH4 concentrations exhausted from underneath a hood designed to partially enclose the volume above a feed bin. The principle is almost similar to GF system except that there is no requirement to offer extra feed supplements needed to influence the visit of cattle into GF system [73]. There are also some other indirect approaches have been suggested and so far used to measure enteric CH4 emissions from animal however, associated with lower accuracy and greater uncertainty in the emission data [18].
Methane mitigation from animal origin is a time demanding issue throughout the world. There are several possible targets and mitigation strategies (Fig. 2) have been taken so far but still lack in sustainability. By 2050, the total CH4 emission from ruminant animals is expected to increase significantly due to the increasing demand for milk and meat of animal origin for a hurriedly growing world population [74]. So, it is highly needed to mitigate CH4 emission from the livestock industry. Here, we summarized important methane mitigation approaches in ruminant productions.
Methane production is not consistent for all animal types and breeds [13]. Olijhoek et al. [75] reported that methane production per kilogram of dry matter intake (DMI) was lower in Holsteins in comparison to Jerseys (30.7 vs. 32.6 L/kg of DMI in case of High RFI and low concentrate group, 21.4 vs. 28.2 in High RFI and high concentrate group, 32.4 vs. 32.5 in Low RFI and high concentrate group and 24.5 vs. 27.9 in Low RFI and low concentrate group). It was also reported that CH4 production from different animals under the same feeding trial reveals significant variation among animals [8]. Pinares-Patino et al. [76] conducted an experiment on grazing sheep where some animals show as high and low CH4 emitters on the basis of CH4 output per unit of feed intake [8]. Some other researches have established that ruminants with low residual feed intake (RFI) emit less CH4 than the animals with high RFI [77]. Likewise, Hegarty et al. [78] stated that CH4 production was lower in low RFI Angus steers than in steers having high RFI (142 vs. 192g CH4/day). There was a positive genetic correlation between RFI and predicted methane emission (PME; g/d) which indicated that the cows having lower RFI have lower PME (estimates ranging from 0.18 to 0.84) [79]. Though the cause is not clear, it might be due to differences in methanogen populations among animals [80]. It is proposing that rumen microbial community varies among animals or breeds depending on individual genetic variations that greatly influence CH4 production [42]. Therefore, the selection of low methane producer might be a possible and sustainable way to mitigate methane emission.
Methane production in ruminants is influenced by the composition of feed. Digestion in the rumen is dependent on the activity of microorganisms, which need energy, nitrogen and minerals [81]. Subsequently, the quality of forage affects the activity of rumen microbes and CH4 production in the rumen. The species, processing, and proportion of forages, and the grain sources of diets also influence CH4 production in ruminants. Methane production tends to decrease as the protein content of feed increases, and increases as the fiber content of feed increases [7,82,83].
High-quality forage, especially young plants, can mitigate CH4 production by shifting the fermentation pathway as this forage contains lower amounts of NDF and higher easily fermentable carbohydrates, leading to a higher digestibility and passage rate [84]. On the other hand, more mature forage encourages a higher CH4 yield mainly owing to an increased C: N ratio, which subsequently decreases the digestibility in ruminants [85]. The CH4 emission can also vary depending on types of forage due to the variation of their chemical composition [86]. Methane production also significantly affected by the processing and preservation of forages [23] such as, chopping or pelleting of forages require less degradation in the rumen due to their smaller particles size as a consequent reduction of CH4 emission per kg of DMI [87]. Likewise, ensiling of forages, partially fermented, can reduce CH4 emission from ruminants [87].
For increasing the production especially in the high producing dairy cattle higher energy supplementation is needed. Only forage is not sufficient to provide the nutrient for these high yielding cattle. So, concentrates must be supplemented with forages with a higher density of nutrients and less fiber. These concentrates contain fewer cell walls and readily fermentable carbohydrates (starch and sugar) and contribute to the production of propionic acid however reducing CH4 production [23]. It was observed in one study that 80% and 90% concentrate supplementation decreased CH4 production, while no noticeable effect was found at 35% or 60% concentrate supplementation [88]. Another study showed that a diet containing 90% concentrate produced extremely low CH4 which represented a loss of only 2%–3% of the gross energy intake [7]. Conversely, a diet with high-concentrate contains low in structural fiber and influence to develop sub-acute or acute acidosis. Therefore, dietary manipulation such as feeding a good combination of F:C ration would be effective with respect to mitigating methane emission without hampering their productivity.
The hydrogen and carbon dioxide or formate are the major substrates for methanogens [89]. And, some microorganisms in the rumen use hydrogen to hydrogenate the double bonds of unsaturated fatty acids. Hence, CH4 production is hampered due to the addition of unsaturated fatty acids to the diet [90,91]. Traditionally addition of fat in the diet has been used in order to increase dietary energy content to meet the energy demand of high-producing dairy cows. Very recent, energy supplementation in a ruminant’s diet is changed from carbohydrate to fat, which contributes to CH4 mitigation. The mechanism of CH4 reduction by fat is due to decreasing organic matter fermentation, fiber digestibility, and thus the methanogenic pathway as well as by the direct inhibition of methanogens in the rumen via the hydrogenation of unsaturated fatty acids [7]. Several other studies also revealed that dietary fat supplementation has potential effects on the reduction of CH4 production from ruminants [92,93]. The unsaturated fatty acids act as an H2 sink within the rumen through dehydrogenation and reduction occurs with the highest rate [87]. It was also reported that supplementation of fat often reduces carbohydrate fermentation due to the toxic effects of fat on cellulolytic bacteria and protozoa, whereas starch fermentation was not affected [92]. Therefore, fat reduces CH4 emission from ruminants.
Ionophores are antimicrobial compounds, for example, monensin, lasalocid and salinomycin, are typically used in beef and dairy cattle production to improve feed efficiency and animal performance [91,94]. They reduce CH4 emission from ruminants significantly. It was reported that monensin supplementation for lactating cows reduced CH4 production [95]. For instance, a 25.6% reduction in CH4 production was recorded with supplementation of monensin to Brahman steers without a reduction in daily gain [96]. Monensin upsurges the acetate and propionate ratio in rumen fermentation through the increasing reducing equivalents which contribute propionate formation in ruminant [10]. Other studies showed that a high dose of monensin reduces CH4 production (g/d) by 4%–10% in dairy and beef cattle [97,98]. Likewise, another report revealed a 30% reduction of CH4 emission in beef cattle fed monensin at 33 mg/kg [99]. Ionophores also hampers survival of protozoa as a consequence the reduction and subsequent recovery of protozoal numbers in the rumen help to CH4 decline up to 30% [99]. However, the inhibitory effects of ionophores on CH4 production may not persist over time, and several microbes already adapted to ionophores [7,10].
Mineral mixture also has effects on enteric methane mitigation. For instance, dietary supplementation of illite feed additive, claysized mineral mixture that contains Mg, Ca, K, Mn, Zn, P, Fe, Al, Si, Co, Se and Mo, at 1% on dry matter (DM) basis has a positive effect on CH4 reduction with increasing VFA production in Han-woo steers [100].
The organic acids such as fumarate and malate, and propionate precursor or substances, are the potential feed additives that mitigate CH4 emission from ruminants when supplemented with feed [97,101–103]. They are supposed to stimulate increased production of propionic acid in the rumen by acting as an H2 sink, in this manner reducing CH4 production [104]. Some other study also reported that organic acids mitigate methane production by up to 17% [103]. The in-vitro study showed that fumarate reduces the CH4 output by 38% in continuous fermenters using forage as a substrate [105]. Conversely, an in-vivo study with growing beef cattle reported CH4 reduction was unaffected by fumarate [106].
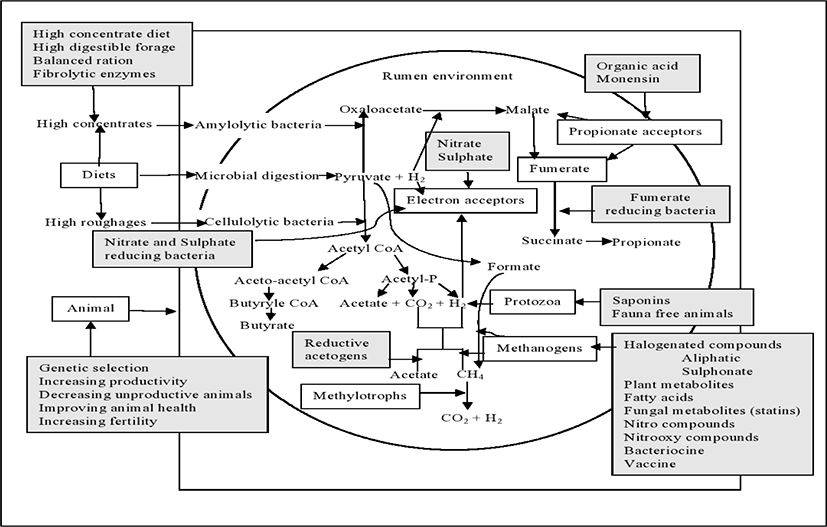
Modification of rumen ecosystem through direct-fed microbials (DFM) or probiotics, is one of the most possible approach to reduce methane production in rumen. Probiotics such as bacterial species including Bacillus, Bifidobacterium, Enterococcus, Lactobacillus, Propionibacterium, Megasphaera elsdeniiand Prevotella bryantii and yeast (Saccharomyces cerevisiae), are used to improve rumen fermentation and feed efficiency [107,108] which could also reduce CH4 emissions from ruminants. Several pieces of research so far have been conducted earlier in order to mitigate CH4 emissions from ruminant with the supplementation of dietary probiotics. Jeyanathan et al. [109] summarized the several rumen biochemical pathways that could be modulated by direct feed microbials to reduce CH4 production from ruminants (Fig. 2). The production of VFAs such as acetic, propionic and butyric acids are mainly depends on the diet offered to the animal. Ruminants produce comparatively more propionate, fed a concentrate-based diet than those fed a high forage diet, which produces more acetate.
Propionate formation, also considered as H2-utilisation pathway, consumes reducing equivalents, pyruvate which is reduced to propionate [110]. Though these H2 are the key precursor for the production of CH4, the increase in propionate formation is proportionally linked with decrease CH4 production. The succinate pathway is the major pathway of propionate production in the rumen where malate, fumarate, and succinate are formed as intermediate products. In this pathway, a mixture of bacteria such as fumarate reducers (e.g. Wolinella succinogenes), succinate producers (Fibrobacter succinogenes), and succinate utilizers (e.g. Selenomonas ruminantium) also involve. The acrylate pathway is an another important propionate producing pathway in the rumen by lactate-utilizing bacteria (Megasphaera elsdenii) [111]. The lactate is required in this pathway, therefore lactate-producing bacteria such as Streptococcus bovis play a regulatory role. There are several studies have been conducted to enhance propionate production in the rumen targeting increase animal productivity through dietary probiotics [112–114] which are indirectly linked with methane mitigation approaches. The decrease in CH4 production was recorded in lactating dairy cows consumed a mixed Propionibacterium jensenii – Lactobacillus spp. direct feed microbials [115]. Mamuad et al. [116] reported that fumarate reducing bacteria alters the rumen microbiome and helping ruminal fermentation, and reducing CH4 production in-vitro. Recently, CH4 production was also reduced through supplementation of fumarate reductase-producing enterococci (Enterococcus faecium SROD) with the increasing of total VFAs in an in-vitro experiment [117].
Nitrate, an alternative H2 sink to CO2 in the rumen, decreases rumen methanogenesis and reduce the toxicity of the intermediate product nitrite during their metabolism. Rumen microbes hastily reduce the nitrate to nitrite, however the reduction rate of nitrite to ammonia is slower. This can cause nitrite accumulation in the rumen [118] and methemoglobinemia in blood. Methemoglobinemia declines the blood’s capacity to transport oxygen to tissues, resulting in low performances or even death of animal in severe cases [119]. The use of nitrate and/or nitrite-reducing bacteria as probiotic is one of the potential solution to avoid this toxicity [120]. The bacteria having the ability to reduce nitrate, nitrite or both compounds are already existing in the rumen, but their concentration is lower than that of their counterpart methanogens [109,121,122]. The main nitrate-reducing bacteria in the rumen are W. succinogenes and S. ruminantium [121,122]. So, it may be beneficial to increase the number and/or the activity of nitrate- and/or nitrite reducing bacteria in the rumen to decrease methanogenesis. Addition of nitrate in the diet increased the number of nitrate reducing bacteria (W. succinogenes and Veillonella parvula) in-vitro [123] but this is may be insufficient to compete with methanogenesis. Thus, giving nitrate and/or nitrite-reducing bacteria as probiotics along with nitrate may progress the nitrate reduction process and subsequently avoid nitrite toxicity. Along with nitrate, Denitrobacterium detoxificans strain NPOH1 decreased 95% [120], and W. succinogenes, S. ruminantium or V. parvula reduced >70% [123] of CH4 production in in-vitro trial. Conversely, there is a lacking on in-vivo data regarding this issue.
The ability of sulphate-reducing bacteria (SRB) to compete with methanogens is largely determined by the introduction of sulphate into the rumen. In anaerobic environments, where sulphate is unlimited, SRB compete with methanogens for common substrates such as H2, formate and acetate. The population of SRB in the rumen is low (105 to 106 cells/mL) and largely from the genus Desulfovibrio and Desulfotomaculum [124,125]. Recently, another SRB belonging to the genus Fusobacterium was isolated from buffalo [126]. Only few studies were conducted on effect of sulphate supplementation alone in rumen methanogenesis [127,128] due to their toxic end product hydrogen sulfide (H2S). Therefore, the sulphate reduction, owing to decrease methanogenesis, may be facilitated by SRB only when sulphate is added as a feed additive. For example, a reduction in CH4 production was recorded in an in-vitro experiment using Fusobacterium sp., as a probiotic with a high sulphate diet where CH4 production at 72h was reduced from 2.66 to 1.64 mmol/g digested dry matter (DM) without H2S accumulation [126].
The reductive acetogenesis is an accepted mechanism of H2 utilization that coexists with methanogenesis in the rumen [129,130]. Acetate, the end product of this reaction, has the additional advantage of being a source of energy for the animal. However, acetogens are less numerous in the rumen environment and less efficient than methanogens in respect of competing for reducing equivalents. Therefore, it is needed to increase the number of acetogenic bacteria in the rumen, which compete with methanogens for hydrogen, as a result of CH4 reduction. Kim et al. [131] and Martin et al. [23] stated that CH4 production was reduced with the dietary supplementation of acetogen probiotics in ruminants.
It is strongly suggested that yeast probiotics possibly stimulates the acetogenic bacteria to compete with methanogens or to co-metabolize H2 as a consequence reducing CH4 formation [81,132]. A 20% reduction in CH4 production was recorded after 48 h of incubation of mixed rumen microorganisms containing alfalfa and a live yeast product [133]. Aspergillus oryzae reduced CH4 production by the reduction of protozoal population (45%) [134]. Therefore, still, there is a big scope to search the more suitable probiotic candidates for the sustainable CH4 mitigation strategy.
Botanical extract or plants secondary metabolites (PSM), viz. saponins, tannins, flavonoids, organosulphur compounds, and essential oils, have potential anti-microbial effects against several types of microorganisms [135]. Several PSMs have been so far identified as a potential agent to reduce CH4 production by methanogens in the rumen [136,137]. Depending on the type, sources, molecular weight, doses, and diet types, the methane reductive capability of PSMs varies significantly. For instance, Joch et al. [138] examined in-vitro methane abetment properties of nine (09) concentrations of α-terpineol, 8-hydroxyquinoline, bornyl acetate, camphor, thymoquinone, α-pinene, and thymol. They reported all compounds tested validated as CH4 reducing agent however effective concentrations varied among individual PSMs. Recently, some see weeds also have potential effects on methane abatement [139] however, need more researches in this regard.
Saponins (triterpenoid saponin, tea saponin, methanol extract saponin) are the potential additives which cause a significant reduction of protozoa population in the rumen, as a result, methane production is reduced [140–142]. Saponins also contribute rumen fermentation, enhance rumen bacterial populations, and ruminant productivity [50,136,137]. Saponins from different sources show various results. Application of Quillaja saponin at 1.2 g/L decreased CH4 production in-vitro, but not at 0.6 g/L [143]. The ivy fruit saponin decreased CH4 production by 40% [144] and saponins from Saponaria officinalis reduced CH4 and abundance of both methanogens and protozoa in-vitro [145]. However, the opposite result was found in other in-vitro studies, where Quillaja saponins at 0.6 g/L did not reduce CH4 production or abundance of methanogen [146,147]. Similarly, Tea saponins (30 g/day) also did not reduce CH4 emission from steers or methanogens abundance [148]. Though, the effects of saponins on methanogenesis and methanogen abundance are greatly inconstant among different studies, still needed to study more.
Tannins (condensed and hydrolysable tannin) also have the potentials to reduce CH4 production from ruminants. Tannins exert their effects by directly inhibiting methanogens as well as indirectly decreasing H2 production as a consequence decreased fiber digestion and the number of protozoa in the rumen [149]. Tannins extracted from Lotus pedunculatus showed inhibitory effects on pure cultures of methanogens [150]. Likewise, inhibition of methanogens in the rumen of goats supplementing tannins as feed additives was reported by Puchala et al. [151]. However, the inconsistent result was found in the case of condensed tannins [152]. Recently, forages with higher levels of tannins, such as clover and other legumes, including trefoil, vetch, sulla and chicory are considering as mitigating agent [153]. For instance, CH4 production was reduced (up to 55%) while ruminants were fed tannin-rich forages [154]. Tannins may exert a similar mechanism like bactericidal or bacteriostatic and inhibit the growth or activity of rumen methanogens and protozoa [155].
Essential oils, another plant secondary metabolites, are volatile components [153] and aromatic lipophilic compounds [156]. It contains the chemical constituents and functional groups such as terpenoids, phenolic and phenols, having potential antimicrobial activities [157], which inhibit the growth and existence of greatest number of microorganisms in the rumen [158]. Due to their lipophilic nature, they have a very high affinity to microbial cell membranes, and at the same time, their functional groups interact with the microbial cell membrane [159]. With the application of essential oil, the methanogenesis decreases especially by reducing microbial populations [160]. The Allium arenarium oil (garlic oil), a highly promising essential oil, was significantly reduced methane production both in-vivo and in-vitro by 12% and 36%, respectively [161].
Enzyme feed additives having fibrolytic activities are used to enhancing fibre digestibility [162], feed conversion efficiency [163], and milk production [163–165] of dairy cows. A reduction in the enteric CH4 production was reported by Arriola et al. [163] where a fibrolytic enzyme additive was supplemented with a lactating cows’ diet (52% dietary forage). Conversely, some other studies revealed that exogenous fibrolytic enzyme additive increased CH4 yield and altered rumen methanogen community composition, without affecting overall density of methanogens [166,167]. Recently, Biswas et al. reported that dietary supplementation of lysozyme enzyme may improve rumen fermentation and reduce CH4 emission in an in-vitro trial [168]. Therefore, more in-vivo study is needed before use of enzymes for methane mitigation strategy.
Defaunation, the removal of protozoa from the rumen, is often linked with an increased microbial protein supply and enhancement of animal productivity. Though protozoa generate a relatively large volume of H2 and formate, and the methanogenic bacteria attach to the surface of ciliated protozoa [169], defaunation is effective to decline CH4 emission. Morgavi et al. [170] revealed that CH4 emission reduced by 20% over a period of 2 years in defaunated sheep. Due to unclear reason, partial defaunation is not effectively reduce CH4 production [171,172]. So far, a variety of techniques have been tested for defaunation experimentally, but none is used routinely due to its’ toxicity problems to the rest of the rumen microbiome as well as the host animals [9]. It has been also reported that immunization or vaccination of sheep with entodinial or mixed protozoal antigens reduced protozoal populations, and produced Immunoglobulin G (IgG) against rumen protozoa [173]. Recently, plant secondary metabolites have been used as potential defaunating agents. Moreover, defaunation technology needs more valuation before use widely and still has a big scope to do more research.
Conclusion
Ruminant production is increasing rapidly for providing good quality meat, milk, and their products to the large population in the earth. Methane production is also rising proportionally that contribute negatively to global warming as a greenhouse gas. Therefore, we should give more concentration on sustainable methane mitigation strategy which could be achievable through a cumulative approach. There are several methane mitigation strategies such as animal intervention, diet selection, dietary feed additives, probiotics, defaunation, supplementation of fats, oils, organic acids, plant secondary metabolites, etc. however, sustainable mitigation strategies are not established yet. We should give more emphasis on biological regulation of methane mitigation through searching of suitable candidates of direct feed microbials. Accurate measurement of methane is also highly needed to make the mitigation approach successful.
















