INTRODUCTION
Milk composition influences not only its nutritional value, but also the income of dairy farms. The dairy industry has focused on improving milk fat content more than any other nutrient content in milk. As consumers’ preference trends change to cheese and the importance of milk protein is being emphasized, milk pricing and studies on the improvement of milk content are focusing more on milk protein. To increase the total milk protein yield from dairy cows, one of the most important factors is to supply a proper quantity of amino acids (AAs) in the animals’ diet. AAs are important not only as the building blocks of protein, but also as biologically active substances [1]. Among AAs, an adequate supply and an understanding of the biological roles of essential AAs (EAAs) are of primary concern. Thus, many studies have focused on the most-limiting AAs, such as methionine and lysine, to investigate the promotional animal products [2,3]. However, there is still a lack of research reported on other EAAs.
When dairy cows received an abomasal infusion of 10 EAAs without phenylalanine (PHE), lower yield of milk protein was observed compared to 10 EAAs infused group [4,5]. In addition, the depletion of valine (VAL) from the diets of dairy cows has been shown to reduce milk protein yield and components, denoting the PHE and VAL are important components of the diets of dairy cows that affect milk protein synthesis. Indeed, the effects of some EAAs on milk protein synthesis and transcriptional function in dairy cows and in bovine mammary epithelial cells are well documented [4,6,7]. However, little has been documented regarding the effects of specific AAs on protein synthesis and the roles of these AAs in cellular signaling in bovine mammary epithelial cells.
EAAs and the cellular energy state are key factors involved in the activation of the mammalian target of rapamycin (mTOR) pathway [1]. The mTOR cascade phosphorylates ribosomal protein S6 kinase 1 (S6K1) and eukaryotic elongation factor 2 (EEF2), eventually increasing β-casein levels [8,9]. However, it is still uncertain whether PHE or VAL regulate protein synthesis and energy-mediated cellular signaling in mammary epithelial level. Therefore, we hypothesized that PHE and VAL might have different effects on protein synthesis-related and energy-mediated cellular signaling. To test this hypothesis, gene transcription and proteomic analyses were used to investigate these effects of PHE and VAL in immortalized bovine mammary epithelial (MAC-T) cells.
MATERIALS AND METHODS
For preparation of treatment samples, each powder of PHE or VAL (Sigma-Aldrich, St. Louis, MO, USA) was added to differentiation medium (DMEM/F12 [Thermo Scientific, South Logan, UT, USA] containing 100 units/mL penicillin/streptomycin [Thermo Scientific], 50 μg/mL gentamycin [Sigma-Aldrich], 5 μg/mL insulin [Sigma-Aldrich], 1 μg/mL hydrocortisone [Sigma-Aldrich], and 5 μg/mL prolactin [Sigma-Aldrich]) with various concentrations; 0.3, 0.6, 0.9, 1.2, and 1.5 mM of PHE or VAL. The vehicle, namely differentiation medium without supplementation of PHE or VAL, was used as a control. The initial concentrations of PHE and VAL in DMEM/F12 were 0.22 mM and 0.45 mM, respectively.
The immortalized bovine mammary epithelial cell line, MAC-T cells (University of Vermont, Burlington, VT, USA), was used in this study. MAC-T cells are functional cells that secrete milk components once differentiated.
MAC-T cells were incubated as previously reposted with minor modifications [10,11]. Briefly, cells were cultured in 10-cm plates at 37°C with 5% CO2 and seeded with growth medium in DMEM/F12 containing 10% fetal bovine serum (Thermo Scientific), 100 units/mL penicillin/streptomycin, 50 μg/mL gentamycin, 5 μg/mL insulin, and 1 μg/mL hydrocortisone. When cells were 80% confluent, the cells were cultured in 6-well plates (5 × 104 cells/well) with a growth medium at 37°C with 5% CO2. The growth medium was then replaced with the treatment medium (aforementioned), and cells were incubated for 72 h. The result cells were harvested for protein quantification, RNA extraction, and proteomic analysis. In addition, supernatants were collected for protein quantification.
Cultured medium samples were centrifuged at 300 ×g for 5 min at 4°C. Adherent cells were washed twice with 1 X PBS (Biosesang, Seongnam, Korea), after which added 200 μL of radioimmunoprecipitation assay buffer (Thermo Scientific) containing Halt™ protease inhibitor cocktail (100×) (Thermo Scientific) was added. After incubating for 10 min at 4°C, cell lysates were harvested and centrifuged at 14,000 ×g for 30 min at 4°C. After centrifugation, the supernatants and cell lysates were transferred to new tubes for protein quantification. The concentrations of protein were measured by Pierce™ BCA protein assay kit (Thermo Scientific) according to a manufacturer’s instructions. A standard curve was generated using bovine serum albumin.
Total RNA was extracted using TRI Reagent (MRC, Cincinnati, OH, USA). The quality and quantity of RNA were determined using a NanoDrop 1000® Spectrophotometer (Thermo Scientific). cDNA synthesis and real-time PCR were performed as described previously, with minor modifications [12]. Briefly, cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad, Seoul, Korea) according to the manufacturer’s instructions. Real-time PCR assays were performed using an AccuPower® 2X GreenStar™ qPCR MasterMix (Bioneer, Seoul, Korea), with cycling conditions consisting of an initial incubation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 55 to 65°C for 30 s, and 72°C for 30 s. The analyzed genes are transporter genes of amino acids and glucose including sodium-dependent neutral amino acid transporter type 2 (ASCT2), large neutral amino acids transporter small subunit 1 (LAT1), and glucose transporter 1 (GLUT1), and protein synthesis-related genes including mTOR, S6K1, and β-casein. β-actin was used as a housekeeping gene. The primer sequences are presented in Table 1.
MAC-T cell lysates treated with 0.9 mM PHE and 0.6 mM VAL were selected for proteomic analysis. To remove salts and detergents, 800 μL of chilled acetone was added to 100 μg of protein and samples were incubated overnight at −20°C. After centrifugation at 14,000 ×g for 15 min at 4°C, supernatants were removed and incubated for 20 min at room temperature to eliminate acetone residue. Cell lysates (100 μg) were then resuspended in 0.1% SDS in 50 mM triethylammonium bicarbonate buffer. Proteins were denatured by adding tris (2-carboxyethyl) phosphine and incubating samples for 30 min at 60°C. Protein alkylation was performed by adding methyl methanethiosulfonate and incubating samples for 30 min at room temperature. Proteolytic digestion was performed by incubating samples with trypsin (protein:enzyme = 50:1, w/w) overnight at 37°C. Prior to analysis by LC-MS/MS, peptides were desalted and concentrated. Total peptides were analyzed by nanoUPLC-mass spectrometry/electrospray ionization quadrupole time-to-flight (nanoUPLC-MS/ESI-Q-TOF; Waters, Manchester, UK). LC-MS/MS analysis was performed using a nanoAcquity system, equipped with a Symmetry C18 (5 μm, 5 mm × 300 μm) precolumn and a CSH C18 (1.7 μm, 25 cm × 75 μm) analytical column (Waters).
Data were analyzed using GLM procedures in SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). Cell viability, concentrations of cell lysates and medium protein and gene expression were analyzed by Duncan’s multiple range test. Each experiment had three replicates. Differences between treatments were considered significant at p < 0.05.
Proteomic analysis was performed for acquisition of 2 analytical replicates for 3 biological sets. The MASCOT search engine version 2.4 (Matrix Science, London, UK) was used for protein identification against an IPI_bovine_database (version 3.73; 30403 entries). False discovery rates were < 0.8%. The emPAI score [13] was used for protein quantification and protein quantity was determined by calculating the relative ratio. The significant protein changes (p < 0.05) were considered cut-off threshold of > 2.0-fold and < 0.5-fold. Gene-ontology analysis was performed using PANTHER (gene list analysis, http://www.pantherdb.org).
RESULTS
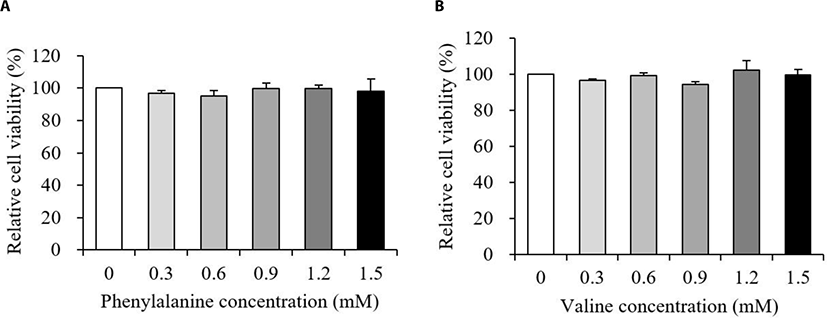
The relative viability of MAC-T cells treated with various concentrations of PHE or VAL is presented in Fig. 1. Results showed that the addition of both PHE and VAL did not affect MAC-T cell viability (p > 0.10).
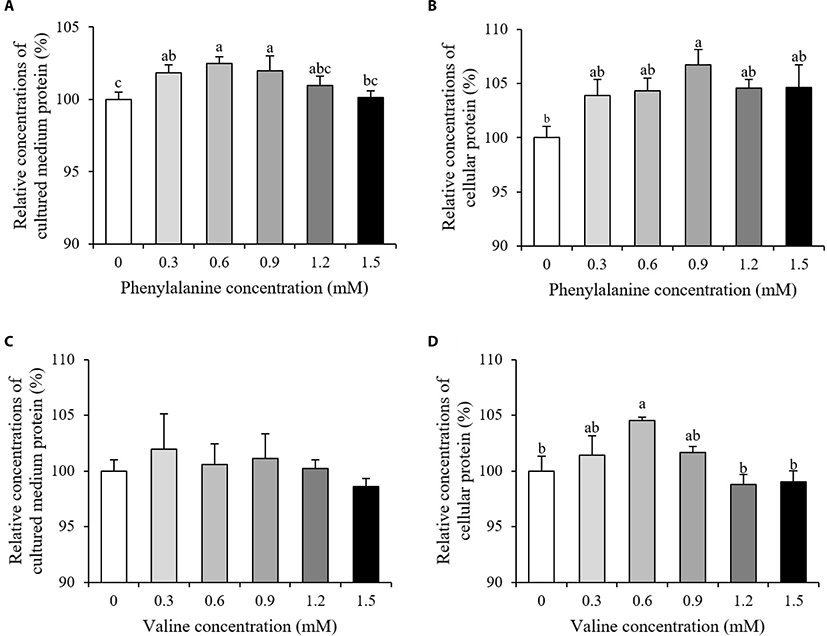
Relative concentrations of cellular protein and protein secreted into the cultured medium are shown in Fig. 2. The relative concentrations of protein in the cultured medium increased in the 0.3–0.9 mM PHE treated groups compared to the control (p < 0.05, Fig. 2A). Of all the PHE treatment groups, only the 0.9 mM group showed an increase in cellular protein (p < 0.05, Fig. 2B). The addition of VAL had no effect on relative concentrations of protein in the cultured medium (p > 0.10, Fig. 2C). However, relative cellular protein concentrations increased in the group treated with 0.6 mM VAL, when compared to the control group (p < 0.05, Fig. 2D).
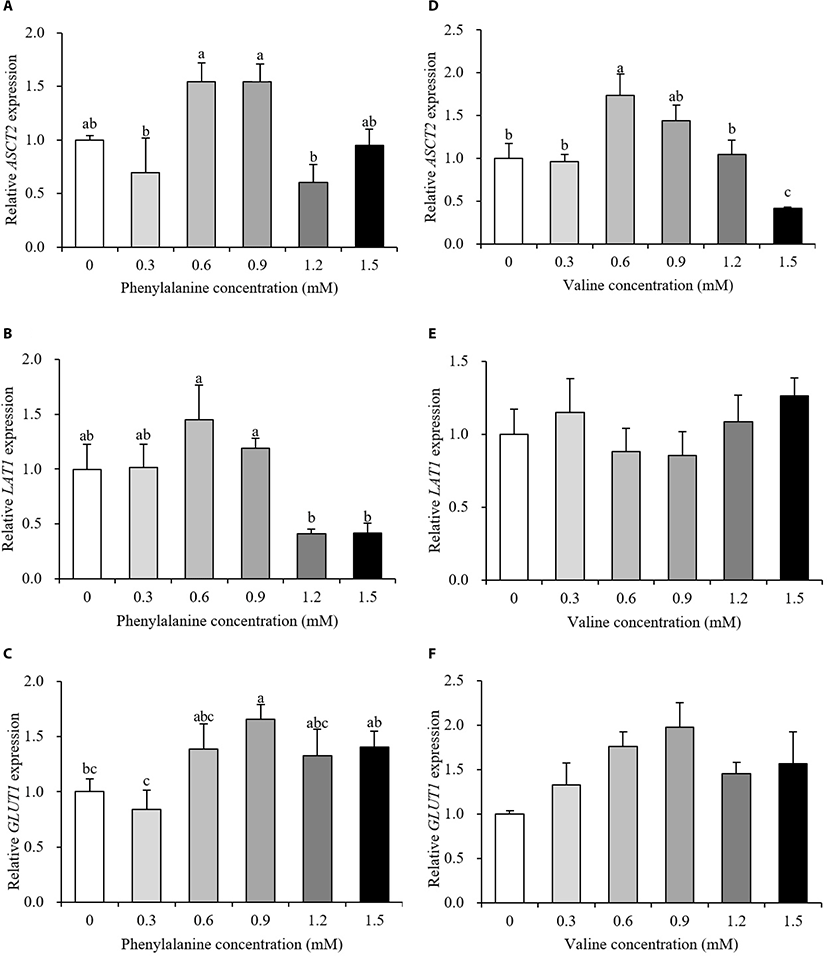
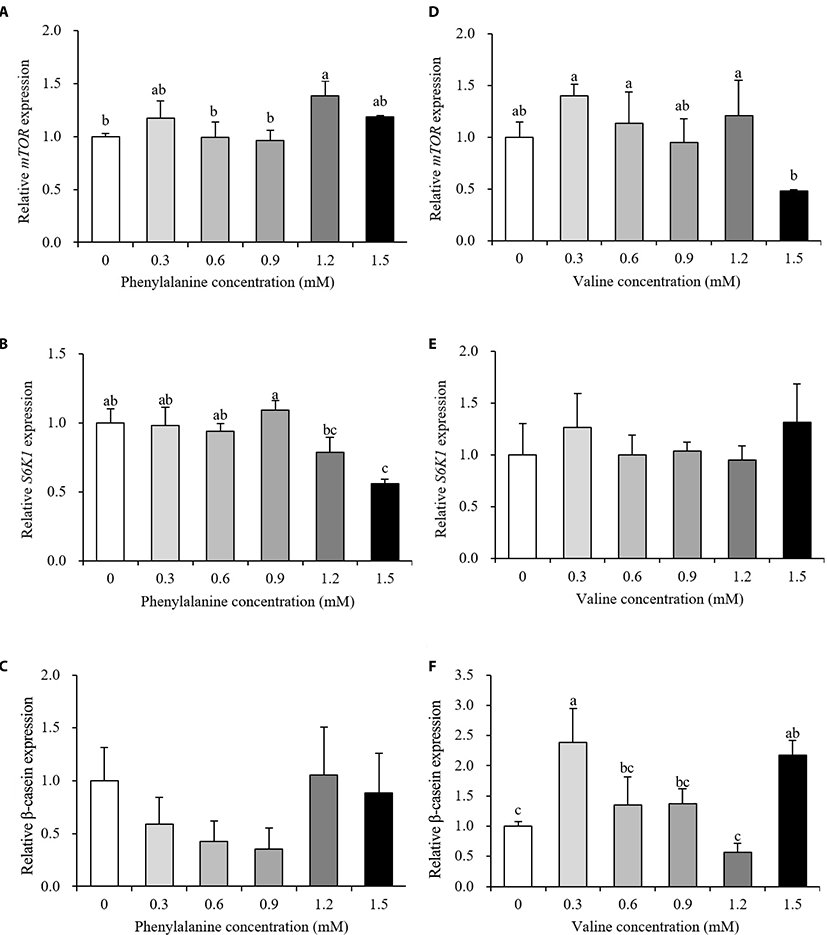
Relative gene expression data are presented in Fig. 3 and 4. The expression levels of transporter genes that transport AAs or glucose from the basolateral membrane to the interior of cells are shown in Fig. 3. After the addition of PHE, ASCT2 and LAT1 expression showed no differences between the control and treatment groups (p > 0.10, Fig. 3A and 3B). PHE treatment, at a concentration of 0.9 mM, increased the expression of GLUT1 by 166% relative to the control group (p < 0.05, Fig. 3C). The expression of ASCT2 was increased by 173% when treated with 0.6 mM VAL (p < 0.05, Fig. 3D). When 1.5 mM PHE was added to the treatment medium, ASCT2 expression was negatively regulated (58% decrease). VAL treatment did not affect LAT1 or GLUT1 expression (p > 0.10, Fig. 3E and 3F). The expression levels of protein synthesis-related genes are presented in Fig. 4. The expression of mTOR increased in the 1.2 mM PHE-treated group by 138% compared to control group (p < 0.05, Fig. 4A). S6K1 expression was down-regulated by 1.5 mM PHE treatment (44% decrease) (p < 0.05, Fig. 4B). The relative expression of β-casein was not changed by the addition of PHE (p > 0.10, Fig. 4C). In the VAL treatment group, neither mTOR nor S6K1 expression was affected (p > 0.10, Fig. 4D and 4E); however, β-casein expression increased after treating cells with 0.3 and 1.5 mM VAL (238% and 218% increase, respectively) (p < 0.05, Fig. 4F).
A total of 134, 142, and 133 proteins were detected in control group, PHE treated group, and VAL treated group, respectively (Table 2). The 61 proteins were up-regulated and 53 proteins were down-regulated after PHE treatment. In VAL treated group, 59 proteins were up-regulated and 63 proteins were down-regulated. Total detected proteins are presented in additional file (Table S1).
Among the fold-changed proteins, that are involved in protein synthesis or energy metabolism are presented in Table 2. In PHE group, ATP synthase F1 subunit beta (ATP5B) was 2.0-fold up-regulated compared to control. Three proteins were detected after PHE treatment, however not detected in control group: eukaryotic translation elongation factor (EEF) 1 alpha 1 like (LOC784131), EEF 1 beta 2 (EEF1B2), and EEF 1 delta (EEF1D). Four proteins were not detected after PHE treatment although detected in control: eukaryotic translation initiation factor 4A 1 (EIF4A1), EEF 1 alpha 1 (EEF1A1), EEF 1 gamma (EEF1G), and EEF2. ATP5B was 3.0-fold up-regulated in VAL treated group. Three VAL treated proteins were detected, but not in control: EEF 1 alpha (EEF1A), EEF1B2, and EEF1D. Three proteins were down-regulated after VAL treatment compared to control: ATP synthase F1 subunit alpha (ATP5A1), 2.2-fold; lactate dehydrogenase A (LDHA), 4.8-fold; and EEF2, 3.0-fold. EIF4A1, EEF1A1, and EEF1G were not detected in VAL, however detected in control.
Biological pathways were analyzed those involving fold-changed proteins in MAC-T cells after PHE or VAL treatment (Table 3). The identified pathways were generally involved in biological regulation, cellular process, and metabolic process. This pathway analysis suggested that PHE and VAL have different biological roles in cells, especially with regard to energy metabolism.
Pathway analysis was performed using PANTHER (http://www.pantherdb.org).
DISCUSSION
The objective of this work was to evaluate the effects of PHE and VAL on milk protein synthesis-related and energy-mediated cellular signaling in vitro using MAC-T cells. We hypothesized different effects of PHE and VAL on cellular signaling. Thus, several analyses were conducted including protein quantities, gene expression, and proteome to test effects of PHE and VAL in MAC-T cell signaling.
Cell viability was analyzed to investigate whether the addition of PHE or VAL had a negative effect on MAC-T cells. In our results, neither PHE nor VAL affected viability of MAC-T cells between 0.3 and 1.5 mM (Fig. 1A and 1B, p > 0.10). Thus, further analyses were performed to investigate the effects of PHE or VAL on protein synthesis-related responses and energy-mediated cellular signaling in MAC-T cells.
When high milk-producing Holstein cows fed a rumen protected form of PHE, little or no change in body conditions score were observed and there is either a tendency towards a decrease or no change in true milk protein yield compared to control group [14–16]. However, our study showed a significant increase in concentrations of protein in both the cultured medium and within cells after PHE treatment (Fig. 2A and 2B, p < 0.05). This can be explained as differences between individual variation of animals and controlled laboratory environment. Results from our study suggested that PHE has the potential to increase both secretory and intracellular protein in mammary epithelial levels. Positive effects of branched-chain amino acids (BCAAs) on both milk and body protein synthesis have previously been documented [17]. In porcine mammary tissue, after the addition of VAL to the culture medium, the amount of intracellular VAL exceeded the amount of extracellular VAL within 15–30 min [18]. In this experiment, only cellular protein concentrations were significantly increased after VAL treatment (Fig. 2C and 2D, p < 0.05). The absorbed VAL in interior of mammary cells may utilized for the intracellular protein synthesis.
In results of milk protein synthesis-related and energy-mediated gene expression, the ASCT2 transports BCAAs, threonine, and some non-essential AAs to the cell interior and LAT1 is involved in the transportation of EAAs except arginine, lysine, and threonine [19]. ASCT2 and LAT1 expression did not change after treatment with PHE compared to control (Fig. 3A and 3B). Our results suggested that the addition of PHE may not regulate the transport of other AAs into cells. Further research is required to understand the regulation of AAs by PHE. VAL is transported by both ASCT2 and LAT1 in the basolateral membrane. Our results showed that addition of VAL affected ASCT2, but not LAT1 (Fig. 3D and 3E). According to Dong et al. [20], LAT1 expression was not affected by additional VAL above the ideal profile of EAAs in MAC-T cell medium. These results demonstrated that VAL is transported through ASCT2. In general, glucose is used in mammary epithelial cells as a substrate for the synthesis of milk components and energy production [21]. Because mammary glands lack glucose-6-phosphate, mammary epithelial cells need to transport glucose to meet their energy requirements. Our results showed the stimulation of GLUT1 gene expression after the addition of 0.9 mM PHE. These results suggested that PHE can contribute to energy metabolism by increasing glucose uptake.
The mTOR is a central protein of the signaling pathway that regulates translation and is involved in protein synthesis [9]. S6K1 is one of the downstream mediators of the mTOR signaling pathway. β-casein is not only a large component of milk protein but is also a final product of the mTOR translation initiation pathway. When bovine mammary epithelial cells were differentiated with AA-free medium supplemented with PHE, the phosphorylation status of mTOR was significantly increased compared to the negative control; however, the phosphorylation of S6K1 is not affected [22]. In the same study, both mTOR and S6K1 levels were shown to increase after VAL treatment. Similar studies have reported that orally administered VAL, but not PHE, increased S6K1 levels in the rat pancreas [23]. Furthermore, when additional VAL was added to MAC-T cell culture medium containing an ideal profile of AA, mTOR levels were significantly increased and S6K1 levels showed an increasing trend [20]. These reports partially agree with the results obtained in the present study reporting an increase in mTOR expression and no change in S6K1 expression after PHE treatment (Fig. 4A and 4B). However, we observed no change in mTOR or S6K1 levels in cells treated with VAL (Fig. 4D and 4E). The PHE treatment group showed either no change or a slight up-regulation of mTOR, no change or a slight down-regulation of S6K1, and no change in β-casein levels. Nevertheless, with no effect on mTOR or S6K1 expression, VAL treatment increased the relative expression of β-casein. These results may be explained by the fact that protein synthesis is a highly complex process that involves energy status, hormones, and nutrients. Taken together, our results of GLUT1 expression suggest that glucose uptake may be positively regulated by PHE and that VAL has the potential to affect AA uptake and may lead to an increase in milk protein yield.
The samples for proteomic analysis were prepared from the 0.9 mM PHE and 0.6 mM VAL treatment groups that were maximized for cellular protein quantities (Fig. 2B and 2D). Interestingly, proteins that are involved in energy metabolism or translation were found to be up- or down-regulated. ATP5A1 and ATP5B are subunits of mitochondrial ATP synthase. ATP5A1 was down-regulated only in the VAL treatment group, but ATP5B increased in both PHE and VAL treatment groups (Table 2). The protein synthesis of mammary epithelial cells requires high levels of ATP [24]. According to Dai et al. [25], the increased abundance of ATP5B was observed when comparing lactating and nonlactating bovine mammary gland. These results are in line with the relative expression levels of GLUT1 observed after 0.9 mM PHE treatment (Fig. 3C) and the detection of ATP synthesis pathway after PHE and VAL treatment (Table 3). Results suggested that PHE has a potential to modulate energy utilization. In the present study, LDHA was especially down-regulated by 4.8-fold after VAL treatment (Table 2). The lactate dehydrogenase (LDH) is an inter-conversional enzyme between pyruvate and lactate, NADH and NAD+ [26]. In four isomer forms of LDH, especially LDHA and LDHB are known to contribute to pyruvate and lactate conversion. LDHA has a high affinity for pyruvate and therefore, it prioritizes the conversion of pyruvate to lactate and NADH to NAD+ [26]. Taken together, down-regulated LDHA in the VAL treatment group indicate that pyruvates had more opportunities for contact with the TCA cycle and therefore, more chances to produce ATP.
Eukaryotic elongation factor has a role in the ribosome-mRNA complex, where it helps continue the growth of the peptide chain. In our proteomic analysis, EEF1A like, EEF1B2, EEF1D were detected in PHE, and EEF1A, EEF1B2, EEF1D were detected in VAL (Table 2). EEF1A helps AA-tRNA complexes attach to the A site of the ribosome and EEF2 then allows ribosomes to read the next codon in the mRNA strand [8,27]. EEF1B, which converts EEF1A-GDP to EEF1A-GTP, is partially bound to valyl tRNA synthetase (ValRS) [27]. Moreover, because cellular VAL increased in response to increased ASCT2 expression (Fig. 3D), ValRS may be activated with the up-regulation of energy-related pathways (Table 3). Subsequently, EEF1B, combined with ValRS, may change EEF1A to an available state, finally resulting in the up-regulation of EEF1A. Although β-casein expression increased in the VAL treatment group in the present study (Fig. 4F), EEF2 decreased. These results may coincide with the lack of effect seen on S6K1 expression (Fig. 4E). S6K1 can phosphorylate EEF2 kinase [8]. However, S6K1 expression was not affected by VAL treatment in the present study. Thus, EEF2 kinase may not have been phosphorylated, denoting that EEF2 was down-regulated. Although EEF2 decreased after VAL treatment, β-casein expression still increased, most likely because protein synthesis requires various factors.
Collectively, these data show that PHE treatment increased both secreted and intracellular protein quantities. VAL increased intracellular protein quantity without affecting cell survival. The relative expression of GLUT1 was up-regulated by PHE and VAL increased ASCT2 expression. PHE increased mTOR mRNA expression. VAL treatment had stimulatory effect on β-casein expression. Proteomic analysis explained the relationship between energy metabolism and mammary cell translation elongation due to PHE and VAL treatment. In conclusion, PHE and VAL affected protein synthesis-related and energy-mediated cellular signaling differentially in immortalized bovine mammary epithelial cells. The current study may help increase our understanding of the milk protein synthesis-related roles of AAs in mammary epithelial levels in vitro.
















