INTRODUCTION
The performance of racing horses is primarily related to their energy metabolism, and numerous enzymes and metabolites are involved in this process [1]. The total muscle blood flow, oxygen consumption, and cardiac output also play key roles in race performance [2]. During racing, horses experience a metabolic stress intricately linked with electrolytic loss and energy metabolism [3]. The energy consumption is mainly dependent on the production of adenosine triphosphate (ATP) in muscle, which generates energy through three mechanisms. The phosphocreatine-ATP system provides instant energy when performing short and high-intensity exercises; the muscle glycolytic system involves anaerobic production of ATP, and is limited when lactate concentration reaches its threshold range; and the oxidative system provides more energy through oxidation of glucose, fatty acids and, proteins [4]. To reveal the role of metabolites in race performance, high-throughput techniques such as nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry are being used [5]. In recent years, NMR-based studies have been used to quantify many metabolites in serum, plasma, urine, and tissues [6,7].
Branched-chain amino acids (BCAAs), e.g., isoleucine, leucine, and valine, play important roles in the skeletal muscle metabolism. These amino acids are essential amino acids which activate protein synthesis after exercise. Supplementation with BCAAs in combination with resistance exercise led to an increase in the phosphorylation of p70 (S6k) in human skeletal muscle [8]. Leucine regulates signaling pathways involved in translational control of protein synthesis in skeletal muscle [9]. Inhibition of AMP-activated protein by leucine stimulates mammalian target of rapamycin (mTOR) signaling in C2C12 myoblasts [10]. The catabolism of BCAAs in skeletal muscle is well studied in human and rats [11,12]. Two enzymes, namely the branched chain (alpha) keto acid dehydrogenase complex (BCKDH) and branched chain (alpha) keto acid dehydrogenase kinase complex (BCKDK), tightly regulate this pathway [12,13]. These enzymes are abundant in the inner mitochondrial membrane in various tissues. The BCKDH multienzyme complex consists of three enzyme units namely E1 (α-ketoacid dehydrogenase), E2 (dihydrolipoyltransacylase), and E3 (dihydrolipoamide dehydrogenase). The E1 subunit consists of E1-α and E1-β chains encoded by BCKDHA and BCKDHB [14,15]. The catalytic activity of this enzyme is further regulated by the BCKDK complex. This complex is encoded by BCKDK. Various mutations and defects in BCKDHB and BCKDK are associated with maple syrup urine disease and neurological defects in human [16,17].
Various studies focused on BCCA concentrations in plasma or serum in the post-exercise period have reported an increased concentration of BCCAs [18]. Most of the studies quantified metabolite levels in endurance racehorses [19,20]. The effects of metabolites in the skeletal muscle have not been studied well, which restricts the analysis of metabolites for muscle physiology and energy metabolism. Moreover, studies evaluating differences between breeds of horses at the metabolomic and gene expression levels have not been reported to date, despite its importance in the analysis of racing performance in horses. The purpose of this study was to compare the metabolite and gene expression levels involved in energy production during the pre- and post-exercise period in two breeds of horses.
MATERIALS AND METHODS
Two stallions and one mare Throughbred horses aged 5, 9, and 10, weighing from 500 to 700 kg which were maintained at Ham-an Racing Horse Resort and Training Center were used to obtain the blood and skeletal muscle samples before and after exercise. Exercise was performed by trotting at the speed of 13 km/h for 30 min and cantering through lunging and long-reining (circular bridge lunging).
Three Jeju horses (3 mares), which were maintained at The National Institute of Subtropical Agriculture, Rural Development Administration were used to obtain tissue samples skeletal muscle, kidney, thyroid, lung, appendix, colon, spinal cord and heart. Venous blood samples were collected using a 20 mL syringe and transferred to ethylenediaminetetraacetic acid (EDTA)-containing tubes. For the skeletal muscle biopsy, local anesthesia was administered to the gluteus medius in muscle, and a biopsy collection syringe was then used to obtain the muscle samples. All samples were stored at –80°C before RNA extraction. All procedures were conducted by following the protocol that had been reviewed and approved by the Institutional Animal Care and Use Committee at Pusan National University (protocol numbers: PNU-2013-0417, PNU-2013-0411, PNU-2015-0864).
The horse muscle-derived cells were established in our previous study [20]. The horse muscle cells were routinely cultured in medium 199 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Invitrogen, Waltham, MA, USA) and 1% antibiotic–antimycotic (Invitrogen), and kept at 37°C and 5% CO2 environment. Routine medium changes were performed three times a week. Cells at 70% to 80% confluency were gently washed twice with phosphate-buffered saline (PBS) and harvested using 0.05% trypsin-EDTA (Welgene, Gyeongsan, Korea) for expansion.
To induce various stresses, horse muscle cells at 70% to 80% confluency were incubated with 20 µg/mL cortisol [21].
Horse muscle-derived cells from the initial culture were plated in a 6-well plate and incubated for 24 h. They were then treated with 20 µg/mL cortisol and incubated for 8 h then harvested. A mixture made of lysis buffer and 2-mercaptoethanol (1 mL:10 µL) was added to the harvested cells, followed by an equivalent volume of 70% ethanol, and the mixture was vortexed thoroughly to ensure complete cell lysis. The mixture was then transferred to the spin cartridge with a collection tube and centrifuged at 12,000×g for 15 s at room temperature. After centrifugation, the flow-through was discarded and the spin cartridge was reinserted into the same collection tube. Then, 700 µL of wash buffer I was added, and the mixture was centrifuged at 12,000×g for 1 min at room temperature. The flow-through was discarded and the spin cartridge was inserted into a new collection tube. After, 500 µL of wash buffer II was added and the mixture was centrifuged at 12,000×g for 1 min. The flow-through was discarded and the spin cartridge reinserted into the same collection tube. This process was repeated and additionally centrifuged at 13,000×g for 1 min to dry the membrane with bound RNA. After, the flow-through was discarded and the spin cartridge inserted into a recovery tube of 1.5 mL. Thirty µL of RNase-free water was added to the center of the spin cartridge and incubated for 1 to 5 min and then centrifuged at 12,000×g for 1 min to elute the RNA from the membrane into the recovery tube. RNA quantity was determined using a spectrophotometer. RNA measurements obtained were then used to calculate the volume of RNA, H2O, 5xBF, dNTP, RNAse inhibitor, OligodT, and RTase needed for cDNA synthesis, and the mixture was subject to reverse transcription.
To quantitate the gene expression levels of BCKDK in muscle tissues and blood cells before and after exercise, a quantitative reverse transcription polymerase chain reaction (qRT-PCR) was conducted using the BioRad CFX-96 apparatus (BioRad, Hercules, CA, USA). Each reaction was conducted in a 25 μL mixture containing 14 μL of SYBR Green Master Mix, 2 μL of forward primer (5 pmol), 2 μL of reverse primer (5 pmol), 5 μL of distilled water, and 2 μL (50 ng/μL) of cDNA. PCR conditions were as follows: a predenaturation step at 94°C for 5 min; 39 cycles at 94°C for 20 s, 56°C for 20 s, and 72°C for 30 s; and a final step at 72°C for 10 min. All measurements were performed in triplicate for all specimens, and the 2−ΔΔCt method was to compare the data [22]. The relative expression level of each target gene was calculated by normalizing the expression level against that of glyceraldehyde-3-phosphate dehydrogenase.
RESULTS
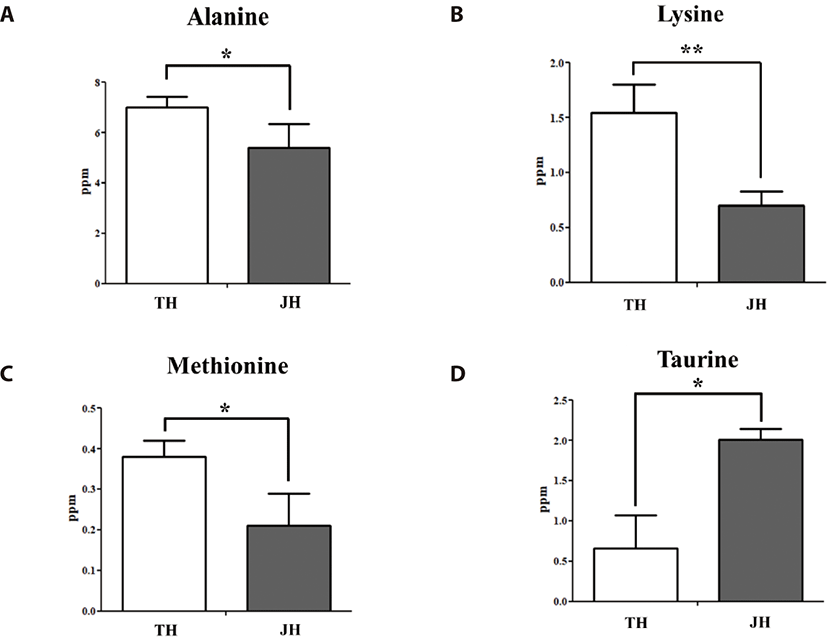
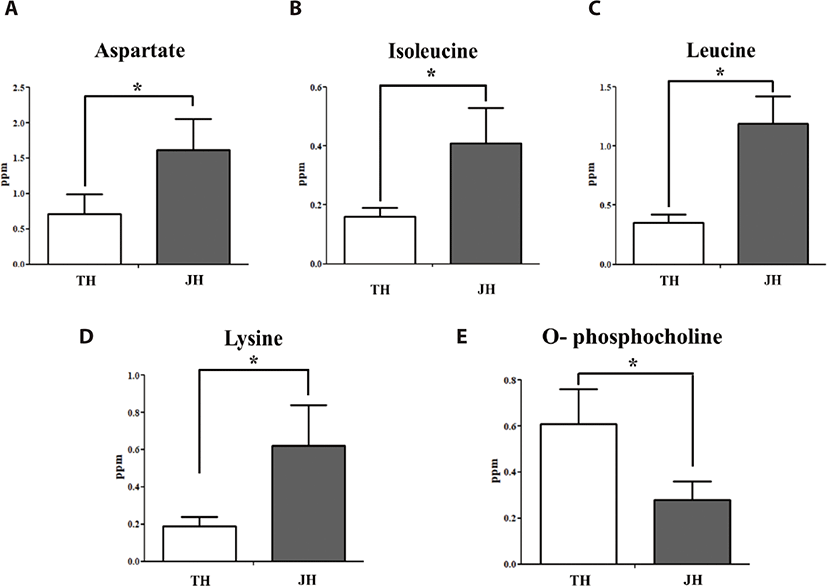
In our previous study, differentially present metabolites were identified in Thoroughbreds [23]. In this study, we identified differentially present metabolites in Jeju horses, and conducted a comparison analysis between Thoroughbred and Jeju horses. We obtained massive metabolomic data from equine plasma (Table 1) and muscle (Table 2). Among the massive metabolites, we obtained each of the four metabolites, which were present at different levels in both the plasma (Table 3) and muscle (Table 3). Alanine, methionine, and taurine were significantly expressed in the plasma sample before exercise, while lysine was significantly expressed after exercise. In muscle samples, aspartate, isoleucine, leucine, and lysine were significantly expressed before exercise, whereas none were significantly expressed after exercise. In addition, we analyzed the levels of metabolites in Thoroughbred and Jeju horses. Jeju horses had a significantly lower level of alanine, lysine, and methionine; and a higher level of taurine in plasma (p < 0.05) than in Thoroughbred horses (Fig. 1). On the other hand, no other metabolites were found to be either significantly low or high in plasma. No significant differences were found between the amino acids and other metabolites after exercise, except for lysine. When compared to Jeju horses, Thoroughbred horses had a significantly higher level of lysine (p < 0.05), (Table 1). The metabolite profile of skeletal muscles in both breeds indicate very few differences at the region of BCCAs and lysine (Table 2). In muscles during the pre-exercise period, Jeju horses had a significantly higher level of aspartate, isoleucine, leucine, and lysine than in Thoroughbred horses (p < 0.05) (Fig. 2). Other metabolites related to exercise did not have a significant difference in skeletal muscle. Thoroughbred horses had a significantly higher level of phospholipid derivative o-phosphocholine than in Jeju horses (p value: <0.05), and no other significant differences were seen for metabolites in muscle (Table 2).
Based on metabolomic data, we found isoleucine and leucine, which are BCAAs significantly expressed in Jeju and Thoroughbred horses. As mentioned, the BCKDH and BCKDK are tightly involved in the BCAA signaling pathway. Therefore, we evaluated BCKDK expression in the muscle tissue of Jeju and Thoroughbred horses.
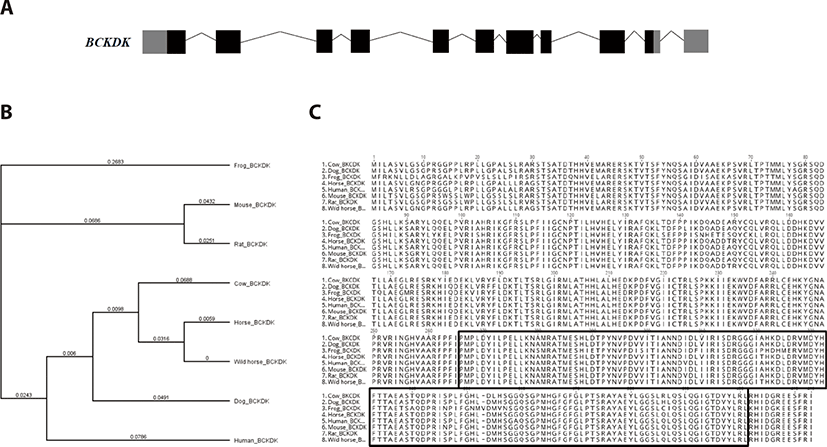
Equine BCKDK is located in chromosome 13, and the genomic structure is shown in Fig. 3. The BCKDK gene consists of 11 exons, and the full lengths is 1,239 bp. Equine BCKDK encodes 412 amino acids. To investigate the evolutionary relationships of BCKDK in horses, we extracted and compared amino acid sequences from eight species (frog, mouse, rat, cow, horse, wild horse, dog, human) from Ensembl 62, and conducted a phylogenetic analysis (Fig. 3B). Multiple alignment using the ‘histidine kinase-like ATPases’ domain showed higher identity (Fig. 3C, solid box). Therefore, we suggest that BCKDK is highly conserved between various species and these domains would have an important role in the exercise stress response.
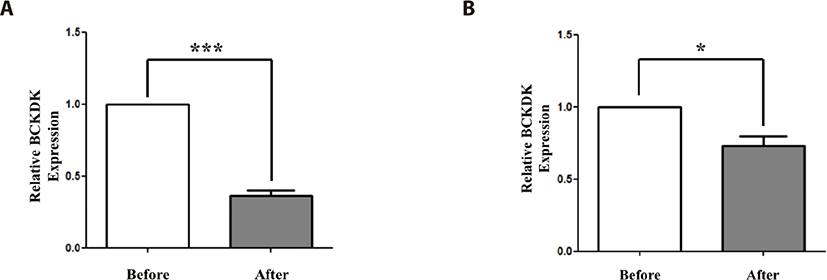
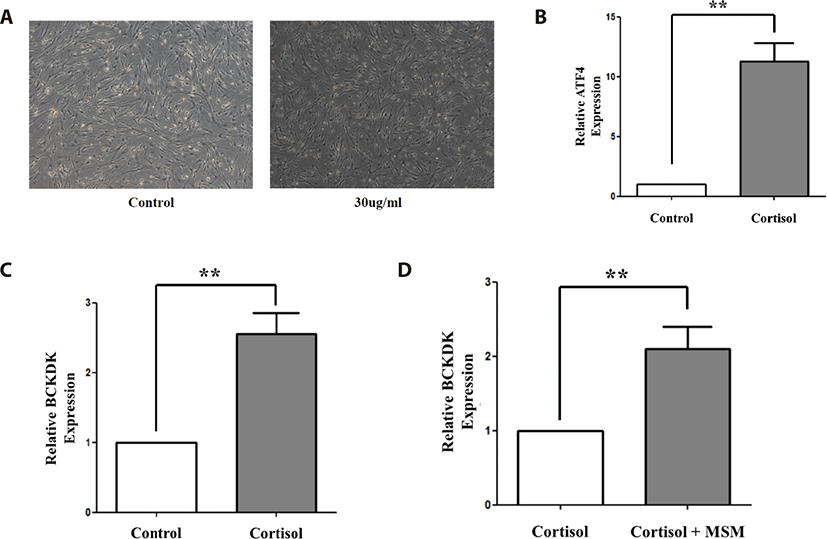
To validate BCKDK gene expression, which we deduced from metabolomic data as a differentially expressed metabolite related gene, we conducted qRT-PCR with Thoroughbred and Jeju horse muscle tissue (Fig. 4). The expression level of BCKDK significantly decreased after exercise in both horse breeds, even though the expression level in Thoroughbred horses decreased more than that of Jeju horses. Additionally, we investigated the expression patterns of BCKDK in horse muscle-derived cells under stress. To validate BCKDK expression under stress, we conducted qRT-PCR on cortisol treated horse muscle-derived cells. In a previous study, we established a cortisol treatment system [21]. In this study, quantitative expression analysis was performed on BCKDK by cortisol reactivity (Fig. 5). To verify the effect of cortisol on stress induction, expression patterns of stress marker genes were investigated (Fig. 5B). We found that the expression levels of the marker genes of stress increased substantially. Next, we examined the effects of cortisol on the horse muscle-derived cells. BCKDK expression level increased after cortisol treatment (p < 0.01, Fig. 5C). In addition, we investigated the effect of methyl sulfonyl methane (MSM) on stress reduction (p < 0.01, Fig. 5D). We found that MSM did not reduce stress by regulating BCKDK. It is assumed that MSM may reduce exercise stress through another signaling pathway.
DISCUSSION
Comparison studies in horses showed that there are variations between amino acid concentrations in skeletal muscle [24]. Among these amino acids, isoleucine and leucine, which are BCAAs, play pivotal roles in exercise physiology [25]. Additionally, the basic amino acid lysine also plays important roles in skeletal muscle metabolism [26]. The comparison analysis shows the physiological status of Thoroughbreds and Jeju horses. Elevated amounts of alanine, methionine, and taurine in plasma pre-exercise in Thoroughbreds, suggest their capacity to perform in races. In skeletal muscle, high amounts of aspartate, isoleucine, leucine, and lysine in Jeju horses indicate their slow ability to respond to exercise. This finding is supported by the low amount of phosphocholine in Jeju horses. In general, alterations in the concentrations of essential amino acids such as methionine, isoleucine, leucine, and lysine in plasma and /or in skeletal muscle reflects the important functions of essential amino acids in moderate exercise. Moreover, various studies on horses showed decreased BCCAs in plasma and changes in skeletal muscle [27]. Generally, exercise induces protein degradation in skeletal muscle, but several studies on humans and horses showed that supplementation of amino acids reduces this process [28]. Furthermore, Thoroughbred horses participate in daily racing practice; this may contribute to the lesser amounts of these amino acids in their skeletal muscle. Thoroughbred horses have been specially bred for sports; and the racing ability of this breed is higher than in Jeju horses [29]. Physiological factors such as body weight and height contribute to racing ability. In contrast to Thoroughbreds, Jeju horses have low weight and height which has been used for mechanical work. A decreased expression level of BCKDK after exercise in Thoroughbred horses indicates their catabolic ability to BCAAs. As a result, low levels of BCKDK enzymes available in skeletal muscle could activate the BCKDH enzyme complex while performing exercise. Despite these results, we propose that low levels of BCKDK in Thoroughbred horses leads to the activation of the BCKDH enzyme complex, and as a result the catabolism of BCAAs is increased in skeletal muscle. These consequent reactions may lead to BCCAs acting as fuels, as well as anabolic signals for protein synthesis in Thoroughbred horses. For Jeju horses, the lack of change in BCKDK gene expression level may lead to continued suppression of the BCKDH complex, which would result in high levels of BCAAs in skeletal muscle. Moreover, binding of the BCKDH complex with BCKDK also plays an important role in this catabolic process. The process has been well studied in rats, and the results have shown that BCKDK capacity to bind to the BCKDH complex crucially affects BCKDH catalytic activity [30]. In this study, we focused on metabolomes and transcriptomes, but not proteomes. Collectively, the results presented indicate that BCKDK genes play important roles in the exercise response.
















