INTRODUCTION
Avian influenza viruses (AIVs) are single-stranded RNA viruses belonging to the Orthomyxoviridae family that infect a variety of birds. Influenza viruses are classified into three types based on their nucleoproteins and matrix proteins (A, B, and C). Type A influenza viruses (H1N1 and H5N1 among others) are the most virulent and have been shown to be the most pathogenic for humans and other mammals [1]. Furthermore, influenza A viruses can be classified as avian influenza (H5N1), swine influenza (H1N1), or other types of animal influenza viruses based on their origin host. The subtype H5 of highly pathogenic avian influenza viruses (HPAIVs) is classified into multiple clades based on the hemagglutinin (HA) protein [2–4]. Depending on their pathogenicity in chickens, they can be divided into two pathotypes. Low-pathogenic AIVs (LPAIVs) are often significantly less virulent, causing mild to severe respiratory disease, as well as a decrease in water or feed consumption and egg production. HPAIVs, however, usually cause fatal infections in chickens [1–4]. HPAIVs are a major economic problem in the poultry industry because of their high mortality rates.
Influenza-induced apoptosis has been observed in a range of cells, including retinal pigment bronchial [5] nasopharyngeal, lung [6,7], and porcine epithelial cells from the intestine and airway [8–10], natural killer cells [11], and human lung ex vivo cultures [12]. Apoptosis can be caused by either direct synthesis of apoptotic mediators or indirect activity of inflammatory mediators and the release of death ligands from infected cells. The influenza virus reportedly triggers apoptosis both in vitro [13,14,15,16] and in vivo [17].
H5N1 virus-induced apoptosis was reportedly delayed in primary human peripheral blood monocyte-derived macrophages, compared to seasonal influenza-infected cells. Given that the intrinsic pathway is responsible for apoptosis [16], the human H5N1 influenza virus enhances production of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and promotes apoptosis in the surrounding cells via cell–cell interaction [18]. H1N1 and H5N1 viruses have been linked to the altered expression of apoptosis-related genes in human lung epithelial cells and mice [19,20].
After infection with the influenza virus, type I interferons (IFNs) are immediately activated and play a pivotal role in inhibiting viral replication and activating innate immune responses by initiating transcription of IFN response genes [21,22]. IFNs are a family of secreted cytokines [23,24] that exert their biological activities by binding specific cell membrane receptors to trigger a well characterized intracellular signaling pathway [25,26] culminating in the transcriptional induction of interferon-stimulated gene (ISGs). Therefore, IFNs generate diverse cellular and physiological outcomes involving antiviral, apoptotic, antiproliferative, antitumor, and immunomodulatory activities through ISGs [26].
Among the ISGs, interferon-alpha inducible protein 6 (IFI6), also known as G1P3, was first identified as an ISG and encodes three splice variants of 130–138 amino acids (~13 kDa) in human [25–27].
IFI6 antagonizes apoptosis in a cellular context. In cancerous cells, IFI6 antagonizes intrinsic apoptosis via IFNs and TRAIL in myeloma cells and via 5-fluorouracil in gastric cancer cells [28,29]. Previous studies reported that IFI6 plays a crucial role in the pathogenesis of diverse malignant diseases, including myeloma and gastric and breast cancers [27–29]. When IFI6 was overexpressed, preservation of mitochondrial membrane potential (ΔΨ) antagonized TRAIL-, IFNs-, and chemotherapeutic drug-induced intrinsic apoptosis. Although our understanding of its biological functions is limited, IFI6 has been characterized as a proliferative and anti-apoptotic factor in cancer cells [27,29]. Unlike in cancerous cells, IFI6 induces apoptosis in virus-infected cells.
IFI6 was found to be an ISG in chickens [26], and its differential expression in the joints of avian reovirus (ARV)-infected chickens was investigated, revealing that it plays a significant role in resistance against ARV infection [30]. In addition, chicken IFI6 (chIFI6) was identified as a differentially expressed ISG in embryos and the bursa of Fabricius of Newcastle disease virus (NDV)-infected chickens [31]. Also, IFI6 in chicken DF-1 cells causes apoptosis and inhibits NDV replication [32]. Nonetheless, with the exception of melanoma differentiation-associated gene 5 (MDA5), signaling mechanisms that regulate the expression of chIFI6 have not been investigated in chickens [33].
The purpose of this study was to examine the molecular properties of chIFI6 and compare the transcriptional profiles of HPAIV-infected chicken tissues, including the lungs, spleens, and tracheas. Furthermore, we investigated whether the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways play a role in the regulation of chIFI6 transcription in DF-1 cells in response to polyinosinic-polycytidylic acid (poly [I:C]; PIC), a synthetic TLR3 ligand.
MATERIALS AND METHODS
Specific pathogen-free White Leghorn chickens (4 weeks old) were purchased from the Poultry Research Centre of the National Institute of Animal Science (NIAS; Hanoi, Vietnam). The chickens had unlimited access to antibiotic-free feed and water. For HPAIV challenge, we used five of 4-week-old White Leghorn chickens per each group, and these chickens received intranasal inoculation with 200 µL of harvested allantois fluid from the infected eggs, containing 1 × 104 50% egg infectious dose (EID50) [30] of A/duck/Vietnam/QB1207/2012 (H5N1), according to the Office International des Épizooties (OIE) guidelines [34]. Tracheal, lung, and spleen tissues were collected from HPAIV- and mock-infected chickens, and stored at –70°C until RNA isolation. All experiments were conducted in compliance with the institutional rules for the care and use of laboratory animals, as well as implementing the protocol approved by the Ministry of Agriculture and Rural Development of Vietnam (TCVN 8402:2010 and TCVN 8400-26:2014).
DF-1 chicken fibroblast cell lines were obtained from the American Type Culture Collection (Rockville, MD, USA) and maintained in the Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS; Biowest, Nuaillé, France). DF-1 cells were cultured at 37°C in 5% CO2 incubator. PIC was purchased from InvivoGen (San Diego, CA, USA), stocked according to the manufacturer’s instructions, and maintained under the same culture conditions as DF-1 cells during PIC treatment.
DF-1 cells were then treated with PIC at doses of 0.1, 1, 5, and 10 µg/mL and incubated for 1, 3, and 6 h, respectively, to check both time- and dose-dependent effects. In addition, DF-1 cells were treated with an NF-κ B inhibitor before PIC treatment and the expression of IFI6 was examined. BAY 11–7085 (BAY, inhibitor of the transcription factor NF-κB) was purchased from Sigma-Aldrich (St. Louis, MO, USA). The inhibitors were treated on DF-1 cells with 5 μM BAY 11–7085, 3 h before treatment with 5 μg/mL PIC. SB203580 (p38 inhibitor), SP600125 (JNK inhibitor), and PD98059 (MEK inhibitor) were purchased from InvivoGen and MedChem Express (Monmouth Junction, NJ, USA), respectively. MEK inhibition was achieved by treating DF-1 cells with 10 μM PD98059 (MEK) for 18 h, followed by 6 h of stimulation with 5 μg/mL PIC. DF-1 cells were treated with 10 μM SB203580 for 1 h to block p38 MAPK, followed by 6 h of stimulation with 5 μg/mL PIC. JNK inhibition was achieved by treating DF-1 cells with 25 μM SP600125 for 1 h, followed by stimulation for 6 h with 5 μg/mL PIC.
Total RNA was isolated from DF-1 cells and chicken tissues using the Pure-link MiniRNA Extraction Kit (Invitrogen, Carlsbad, CA, USA). For quantitative reverse transcription polymerase chain reaction (qRT-PCR), 1 µg of total RNA was used for cDNA synthesis with a ReverTra Ace-α first strand cDNA Synthesis Kit (Toyobo, Osaka, Japan). Sequence-specific primers (Table 1) were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). qRT-PCR was performed using the CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) and SYBR Green (Bio-Rad). Non-template wells without cDNA were used as negative controls. Each sample was tested in triplicates. The PCR conditions were 95°C for 3 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s using a melting curve program (increasing the temperature from 65°C to 95°C at a rate of 0.5°C per 5 s and continuous fluorescence measurement). The qRT-PCR data were normalized relative to the expression of GAPDH and calculated using the 2-∆∆Ct method, where ∆∆Ct = (Ct of the target gene – Ct of GAPDH) treatment – (Ct of the target gene – Ct of GAPDH) control [35].
The amino acid sequences of IFI6 from various species, that is, cow (XP_010800843.1), humans (XP_024301975.1), horses (XP_023490948.1), pig (XP_020951317.1), cat (XP_019692026.1), dog (XP_535344.1), duck (XP_005027772.1), and chicken (NP_001001296.1) were retrieved from NCBI. Amino acids were then aligned using multiple sequence comparisons by log-expectation (MUSCLE) (http://www.ebi.ac.uk/Tools/msa/muscle/). Phylogenetic analysis was performed using the neighbor-joining method [21] with pairwise deletion, 1000 bootstrap replications, and Kimura 2, as described previously [22].
Both t-tests and analysis of variance (ANOVA) statistical tests were conducted to determine the significance levels. Data are shown as the mean ± standard deviation. Duncan’s multiple range tests followed by one-way ANOVA were used for comparison among different incubation times in each group.
RESULTS
Chicken IFI6(ENSGALG00000013575), duck IFI6(ENSAPLP00000010877), and quail IFI6(ENSCJPG00005007073) genes were found on chromosome 2 in chickens, ducks, and quail, respectively. Both chicken and duck IFI6genes have three exons, and complementary DNA sequences (cDNA) of chicken, duck, and quail IFI6genes are 1638, 495, and 324 base pairs, respectively (https://asia.ensembl.org/index.html). The IFI6 transcripts from chickens ( ENSGALT00000022096.4), ducks (ENSAPLT00000011595.2) and quails (ENSCJPT00005011937.1) encode 107 amino acids. Nucleotide sequence alignment revealed that chicken, duck, and quail IFI6genes shared 93.52% (chicken vs quail), 81.79% (duck vs chicken), 82.41% (duck vs quail) of nucleotide identity (Fig. 1A).
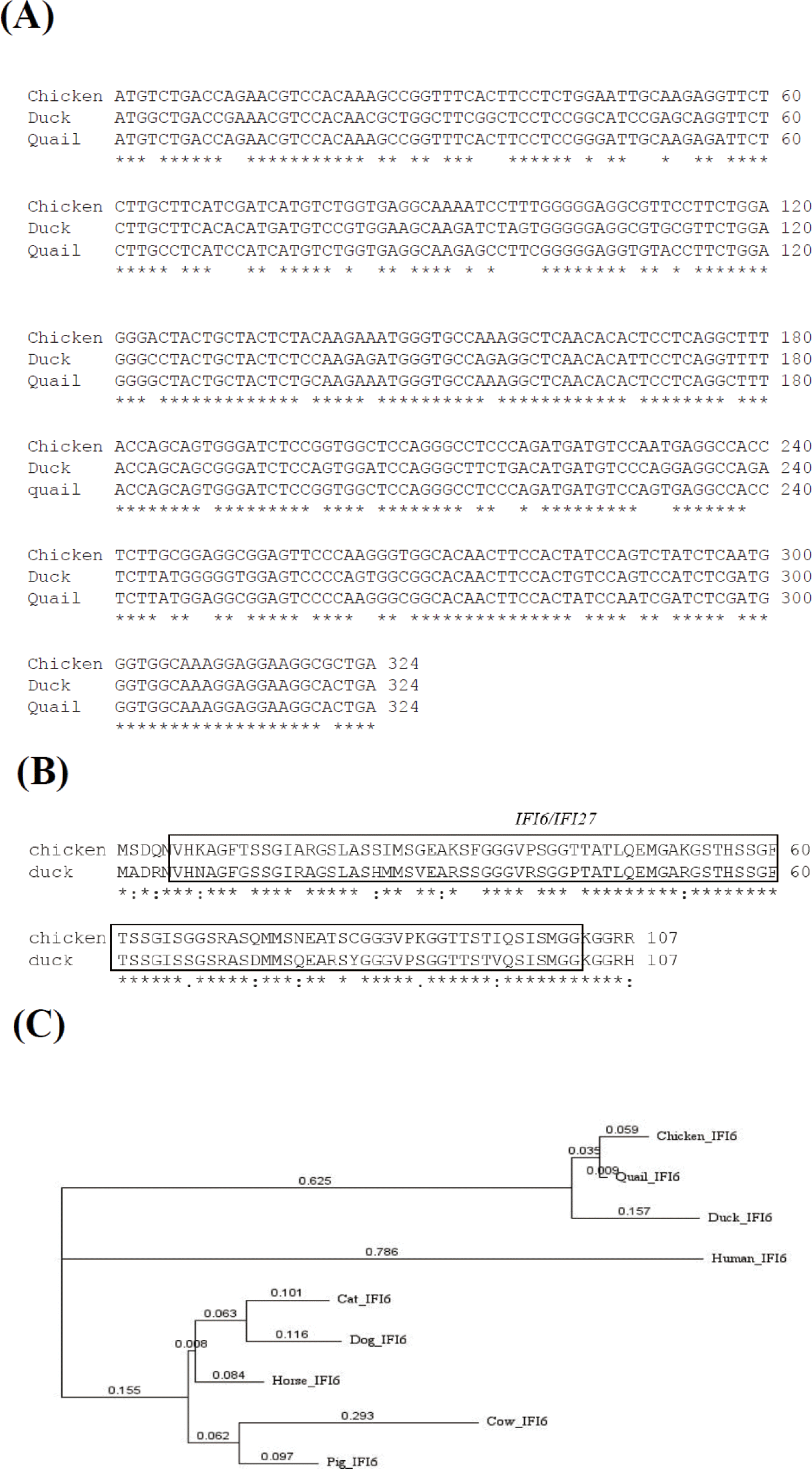
Amino acid sequence alignment revealed that chicken, duck, and quail IFI6 proteins shared 93.46% (chicken vs quail), 79.44% (duck vs chicken), 80.37% (duck vs quail) of nucleotide identity (Fig. 1B), respectively, showing that an IFI6/IFI27-like superfamily domain is conserved in chicken and duck IFI6 proteins (Fig. 1B). To investigate the evolutionary relationships of the chIFI6gene, we obtained cDNA sequences from nine species of vertebrates (chicken, duck, quail, human, cow, dog, horse, cat, and pig) from Ensembl 107 (https://asia.ensembl.org/index.html), and conducted a phylogenetic analysis (Fig. 1C). The results showed that chIFI6 is clustered in the same clade as quail and duck.
We retrieved single nucleotide variants of the chIFI6gene from the Ensembl genome browser.
As a result, 378 variants were retrieved, with six of them being identified in the exonic region.
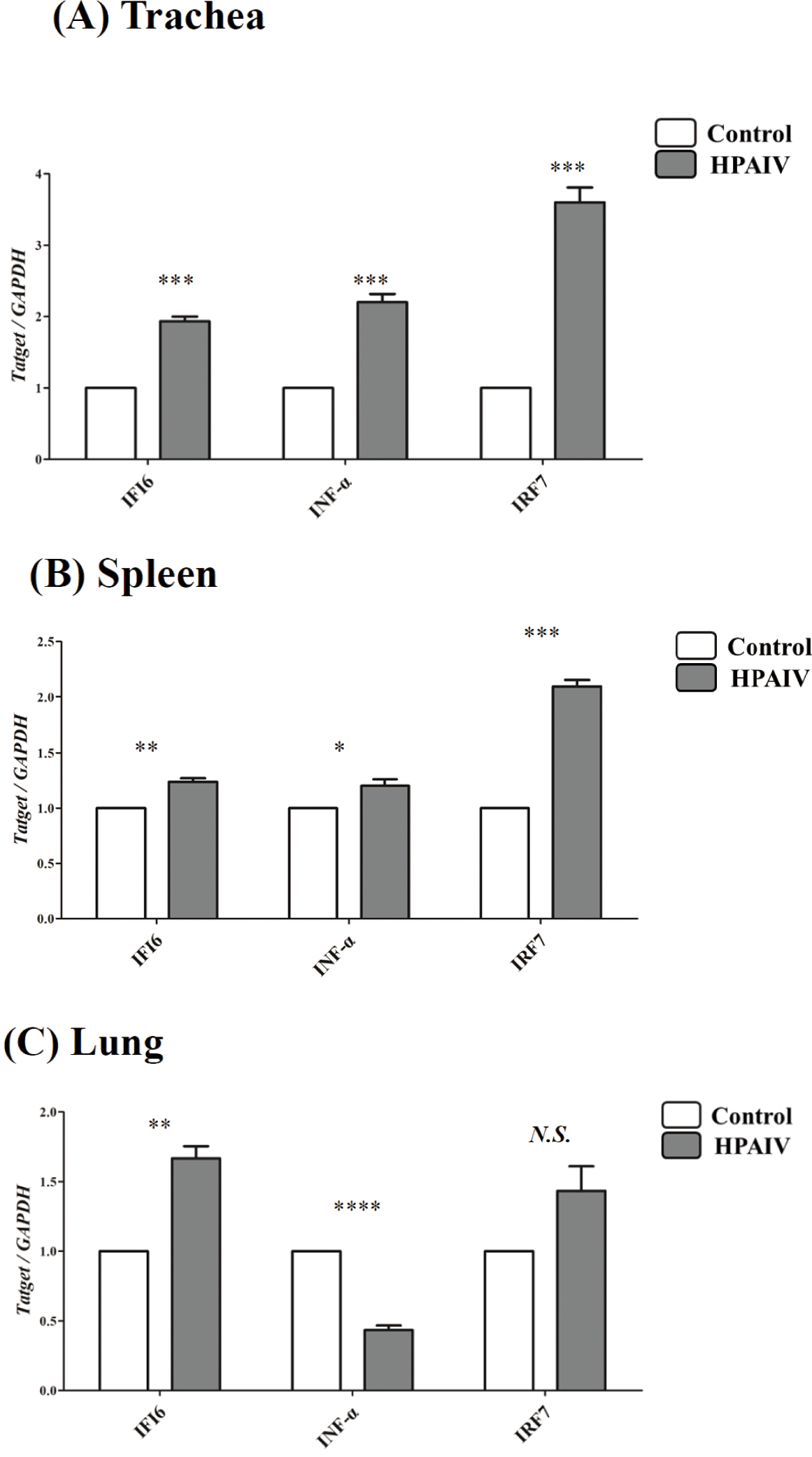
To confirm the differential expression of IFI6during HPAIV infection, qPCR was conducted in the lungs, spleens, and tracheas of HPAIV-infected chickens. The expression of IFI6was considerably higher in the tracheas (p < 0.001, Fig. 2A), spleen (p < 0.005, Fig. 2B), and lung (p < 0.005, Fig. 2C) of HPAIV-infected chickens than in control chickens. In addition, we analyzed the expression of HPAIV infection-related genes, IRF7and IFN-α which are known to be regulated by HPAIV infections. The expression of IRF7, a transcription factor that mediates TLR3 signaling in the nucleus, was dramatically elevated in the spleens and tracheas (p <0.001, Figs. 2A and 2B) of HPAIV-infected chickens, but not in the lungs (N.S., Fig. 2C). Notably, IFN-α expression was significantly elevated in the tracheas of HPAIV-infected chickens (p < 0.001, Fig. 2A), but decreased significantly in the lungs (p < 0.0001, Fig. 2C).
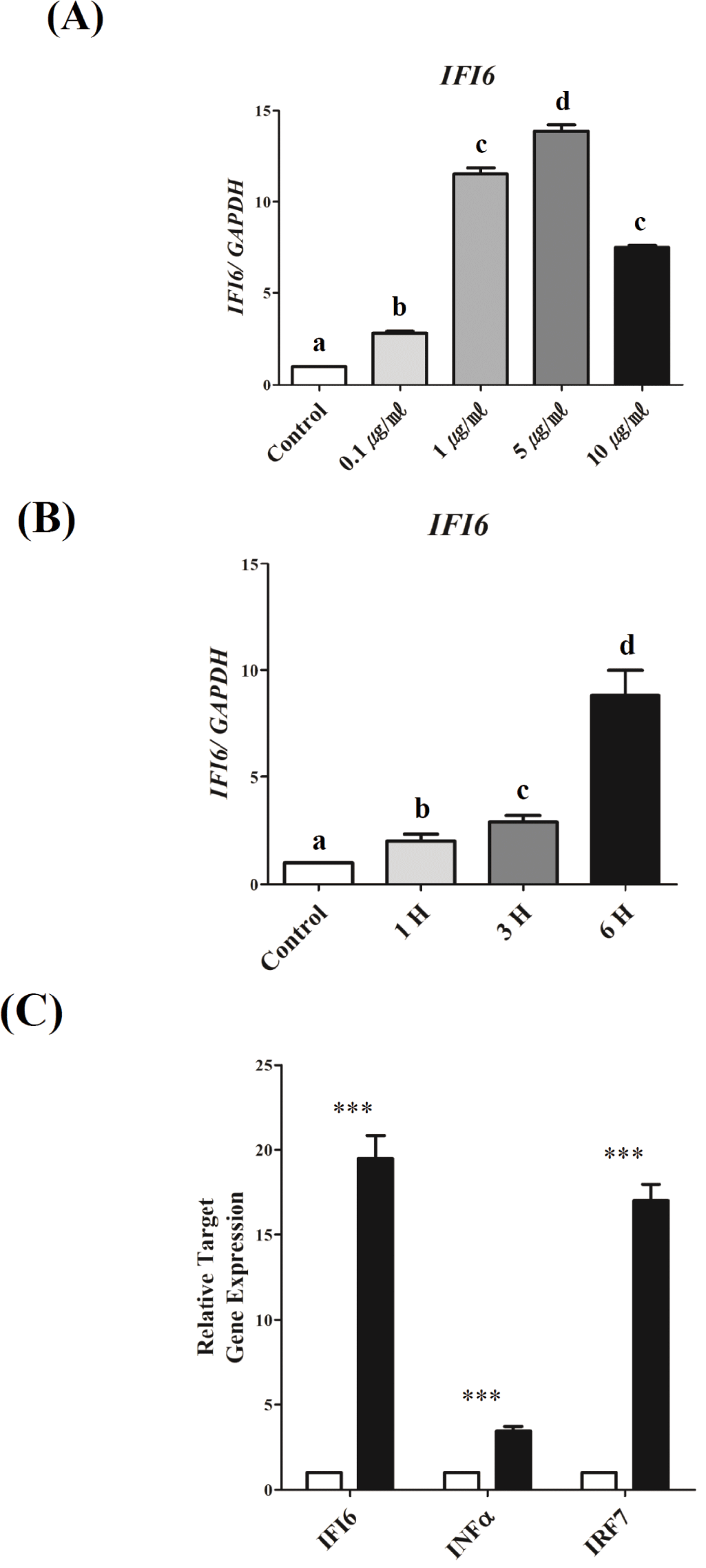
To investigate the mechanisms underlying the transcriptional regulation of IFI6during TLR3-induced inflammation, we used DF-1 cells activated by PIC, as previously reported [36]. IFI6transcription was examined in a dose- and time-dependent manner to establish optimal conditions (Fig. 3). To determine the optimum dose, PIC doses of 0.1, 1, 5, and 10 μg/mL were tested. As a result, IFI6expression was significantly upregulated as the dose of PIC increased from 0.1 to 5 μg/mL (p < 0.001). However, IFI6expression was reduced with 10 μg/mL (p < 0.001, Fig. 3A). To determine the optimum treatment time, DF-1 cells were treated with 5 μg/mL PIC for 1, 3, and 6 h. IFI6 expression was considerably increased depending on the treatment time (p < 0.01, Fig. 3B). Furthermore, the expression of IFNα and IRF7significantly increased under these conditions (p < 0.0001, Fig. 3C).
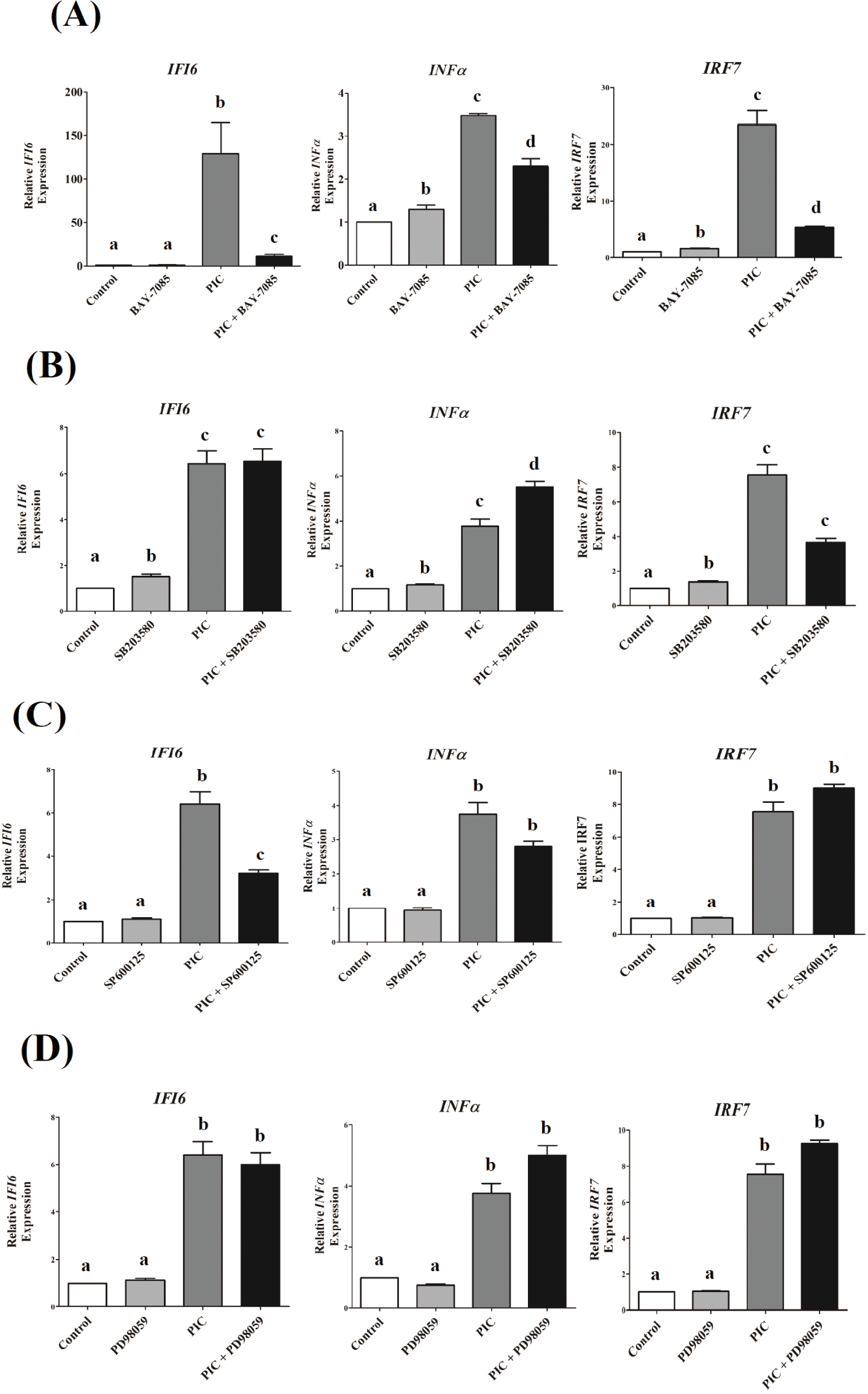
To explore the role of NF-κB and MAPK signaling pathways in IFI6transcription in DF-1 cells that could be triggered by TLR3 stimulation, we used specific pharmacological inhibitors to block these pathways. TLR3-induced transcriptional activation of IFI6was greatly reduced by NF-κB and JNK inhibitors, but not by ERK and p38 inhibitors (Fig. 4). Inhibition of NF-κB and p38 MAPK pathways reduced IRF7expression, which was induced by TLR3activation (p <0.001, Figs. 4A and 4B), but not ERK and JNK (p < 0.1, Figs. 4C and 4D). TLR3-induced INF-α transcription was decreased by inhibiting the NF-κB pathway (p < 0.01, Fig. 4A), but not by inhibiting the MAPK pathways tested in this study ( p > 0.05, Figs. 4B, 4C, and 4D).
DISCUSSION
In this study, we examined the molecular properties of chIFI6, such as nucleotide and amino acid sequence similarity, protein structure, and transcriptional patterns in HPAIV-infected lungs, spleens, and tracheas, as well as transcriptional regulation in chicken DF-1 cells in response to TLR3signalling.
Transcriptional profile analyses revealed that IFI6expression was upregulated in HPAIV-infected lungs, spleens, and tracheas, suggesting a role for IFI6 in pathogenesis, that includes apoptosis, caused by HPAIV infection.
Viruses are intracellular pathogens that can replicate within the cells of living hosts. Consequently, host systems for detecting viral infections and preventing viral replication have emerged. The antiviral response elicited by viral infection is multifaceted and involves the establishment of an antiviral transcriptional program including the synthesis of IFNs, cytokines, chemokines, and the activation of cell death pathways (apoptosis, necroptosis, and pyroptosis) [37]. Individually, these reactions provide notable benefits to the host during viral infections. Type I IFNs can limit viral replication by enhancing the expression of ISGs, which act against the viral life cycle [38]. Thus, ISGs induced by IFNs limit viral propagation in infected cells while promoting an antiviral state in uninfected cells in the surrounding environment [39]. Influenza viruses either inhibit apoptosis in infected cells to use the host cellular machinery for survival and safe replication [40,41], or accelerate cell death to achieve effective replication and transmission, resulting in morbidity [42,43]. Inhibition of cell death by influenza virus A infection is mediated by activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway through direct binding of viral nonstructural protein 1 (NS1) to PI3K, resulting in inhibition of the activity of caspase 9 and glycogen synthase kinase-3 beta [41]. These findings also revealed that PI3K activation confers virus-supporting activity during intermediate stages of the infection cycle; however, influenza viruses can cause massive host cell death in order to replicate and transmit effectively, resulting in morbidity, pathogenicity, and virulence [42,43].
We investigated molecular features of IFI6 in chicken, duck, and quail using genomics data from Ensembl107. As a result, we discovered that nucleotide and amino acid sequences are highly conserved among the avian species studied in this study. A IF6/IFI27 domain, which is unique to FAM14 family member proteins, is also conserved. These findings suggest that the role of IFI6 in infectious disease pathogenesis, as well as the signaling pathway that regulates IFI6transcription, may be conserved, at least in the avian species studied in this study. More research is needed to uncover IFI6’s conserved function and its transcriptional regulation by signaling pathways such as NF-κB and MAPK, and other. In addition, variants of the chIFI6 gene from various chicken breeds that are resistant or susceptible to HPAIV infection will aid in the development of reliable molecular markers for molecular breeding or genomic selection.
In this study, we also discovered that both HPAIV infection and TLR3activation increased IFI6expression (Figs. 2 and 3C). Previous studies have found that viral infections regulate IFI6expression and play a role in viral pathogenesis [44,45]. IFI6expression increased in response to infectious bursal disease virus (IBCV) infection in a previous study [44]. The expression of IFI6induced by dengue virus (DENV) has been previously studied [45]. In another study, IFI6, an ER-localized integral membrane effector, was shown to prevent virus-induced ER membrane formation by controlling some flavivirus infections [46]. Furthermore, a previous study on influenza A virus found that the virus increases IFI6expression in infected cells at 3–18 h timepoints [47]. Nonetheless, the role of IFI6 as an ISG in the pathogenesis of HPAIV infection requires further investigation. Notably, IFI6 overexpression promoted cell apoptosis via a mitochondria-dependent pathway and inhibited in vitroreplication of NDV [32]. In the same study [32], IFI6 protein was found to be localized in the mitochondria, whereas Bax, a pro-apoptotic protein that causes irreversible loss of mitochondrial function, was found to be localized in the cytoplasm. Transcriptional analysis has revealed that genes encoding pro-apoptotic factors (Bax, Bak, Cyt c, caspase-3, and caspase-9) were significantly upregulated in cells overexpressing IFI6, whereas those encoding the anti-apoptotic markers Bcl-2 and Bcl-xl were significantly downregulated [32].
In a previous study, we performed comparative gene expression analyses in PIC-stimulated DF-1 cells [36], which demonstrated that, in chicken DF-1 cells, PIC treatment induces TLR3 signaling cascades to control the target genes from TLRs to proinflammatory transcription factors, cytokines, and type I interferon genes [36]. In these cells, the detection of double-stranded RNAs as ligands triggers various signaling cascades from the endosome to the nucleus, controlling the expression of the target gene. The NF-κB, MAPK, and IRF pathways are among these triggered signaling cascades. In this study, we investigated the transcriptional regulation of chIFI6in DF-1 cells, which was specifically inhibited by NF-κB and MAPK inhibitors. We discovered that NF-κB and JNK were required for TLR3-mediated transcriptional regulation of the chIFI6, whereas ERK and p38 MAPKs were not essential (Fig. 4). TLR3-induced IFN-α expression was unaffected by ERK, JNK, or p38 MAPK inhibition, but influenced by NF-κB inhibition, suggesting that the NF-κB pathway is essential for regulating the transcription of this gene by TLR3 signaling. TLR3-mediated transcriptional regulation of IRF7 is inhibited by suppression of NF-κB and p38 MAPK. These findings imply that the NF-κB pathway is required for the transcriptional regulation of IFI6, IFN-α, and IRF7in DF-1 cells, and that MAPK pathways play a different role in the transcriptional regulation of the genes investigated in this study. Further research into the molecular mechanisms underlying the transcriptional control of chIFI6is warranted.
















