INTRODUCTION
Semen cryopreservation is a major technique for preserving male genetic resources for prolonged periods, as routinely conducted in cattle breeding [1]. In the swine industry, a variety of studies conducted to verify conditions suitable for the practical and economic use of frozen semen. However, Farrowing rates dropped by 20% to 30%, and litter sizes decreased by two to three piglets when artificial insemination (AI) was performed using cryopreserved boar semen [2]. Around half of the semen do not survive cryopreservation due to the sublethal damage, and even when identical amounts of motile semen were utilized for AI, fertility was still lower after thawing than with fresh control semen [3]. Thus, the cryopreservation of boar semen was inadequately used in commercial swine AI programs [4,5], whereas the addition of seminal plasma to the freezing extenders for boar semen drastically improved both farrowing rate and litter size after AI using cryopreserved semen [6,7], enabling successful commercial use. Semen quality after liquid storage or cryopreservation is often analyzed based on motility and morphological features using a computer-assisted sperm analysis (CASA) system [8]. However, the CASA data are insufficient to predict the molecular function of fresh and preserved semen.
Many researches have concentrated on comprehending the process causing cryodamage in order to improve the technique for cryopreserving boar semen. In swine, the sperm that survives the freeze-thawing process is more damaged than the sperm of other species [8]. The reasons for this likely involve the substantial amount of boar ejaculate and the weak semen membrane [8]. Boar sperm are very vulnerable to cold temperatures due to the specific composition of phospholipids in the membrane, which differs from that in other animal species [9]. In addition, boar sperm has exceptional sensitivity to cold shocks because their plasma membrane is rich in unsaturated phospholipids and devoid of cholesterol molecules [9]. As a result of these characteristics, the semen membrane to undergo alterations during chilling and freeze-thawing processes that cause it to become unstable thereby impairing acrosome integrity, calcium homeostasis, and generating membrane lipid disorder [10]. During cryopreservation, because low temperatures modify diffusion and osmosis across this structure, changing the chemical, physical, and electrical characteristics of the cell, a variety of cellular activities that are controlled by the plasma membrane take place [11]. This cell injury is referred to as cold shock. When the plasma membrane becomes unstable at temperatures below 5°C, it can have a severe impact on Ca2+ homeostasis, membrane lipid disorder, and acrosome integrity. Many proteins undergo functional modifications during cryopreservation, including semen metabolism-related enzymes, proteins involved in capacitation and acrosome reaction, proteins related to membrane and structure, and proteins related to apoptosis [12]. All of these changes have an impact on the sperm’s biological functioning, structural integrity, and potential for fertilization. To investigate the mechanism of cryodamage, two-dimensional gel electrophoresis has been utilized to identify alterations in proteins in semen in order to investigate the process of cryodamage, and several differently expressed proteins have been discovered in human [13], sea bass [14], sheep [15], chicken [16], and carp semen [17] after cryopreservation. However, it is still unknown how boar semen changed proteomically after being cryopreserved. At present, in proteomic research, the methods used for quantifying proteins have developed into a combination of isobaric tags for relative and absolute quantification (iTRAQ) and liquid chromatography tandem-mass spectrometry (LC-MS/MS) [18]. Proteomics is a potent analytical method for identifying distinct proteins and elucidating the molecular and biophysical decentralised animal quality control. More accurate expression patterns of boar sperm proteins may be analyzed using a protein chip method as iTRAQ [18], to understand the mechanism underlying the cryodamage of boar semen. Unfortunately, information on the proteome of cryopreserved boar semen is currently very limited [19]. Therefore, in the present study, the iTRAQ 8-Plex labeling system was employed to identify sperm proteins differentially expressed between fresh and frozen-thawed Duroc boar semen.
The purpose of this study was to investigate changes in the proteins of fresh and frozen-thawed semen to understand the primary mechanism and process of cryodamage in semen. Analysis of sperm protein alterations at the proteome level during freeze-thawing offers several potential applications and research value.
MATERIALS AND METHODS
Each ejaculate with a sperm-rich fraction was collected manually using the gloved-hand method from mature 5 Duroc stud boars (14.4 ± 0.8 months old). The experiment was performed in two replicates. Gelatinous material contained in the collected semen samples was removed using 100-μm semen filter paper and the semen samples were diluted with an equal volume (1:1 ratio v/v) of isothermal Beltsville Thawing Solution (BTS; Kruuse, Langeskov, Denmark) at ambient temperature. The diluted semen samples were brought to the laboratory at 25°C within 30 min, and then the quality of ejaculates was evaluated through CASA (ISASv1; Proiser, Valencia, Spain) as sperm motility > 80% and deformation ratio < 15%. The diluted semen samples were split into two parts, one of which was utilized for the cryopreservation analysis, while the other was used for a pooled sample made up of an equal amount of sperm from all animals. The semen collection procedure was approved and implemented in accordance with the standards established by the National Institute of Animal Science, Rural Development Administration (Republic of Korea).
The semen was freeze-thawed with several modifications according to the method described by Westendorf et al. [20]. Diluted semen samples were kept for 2 h at 15°C and then centrifuged at 800×g for 10 min at 17°C. After get rid of the supernatant, the semen pellet was suspended with lactose-egg yolk extender consisting of 11% (v/v) α-lactose and 20% hen’s egg yolk (LEY) to give a final concentration of 1.5 × 109 sperm cells/ml. After stirring slowly, the semen was maintained in a refrigerator (5°C) to further cool for 90 min. The samples in pre-cooled straw racks were transferred into a 5°C chamber. The samples were cooled at −6℃/min from +5°C to −6°C, held for 1 min for crystallization, and cooled thereafter at −40°C/min from −6°C to −80°C and at −60°C/min from −80°C to −150°C, before being plunged into −196°C liquid nitrogen for storage. The straws were defrosted in a water bath with moving water at 37°C for 30 secs. After performing a quality check on the frozen-thawed semen to make sure that each straw had sperm motility that above 30%, the validated semen samples were combined for further analysis.
Fresh and frozen-thawed semen samples were centrifuged to remove supernatant containing seminal plasma and cryoprotectants at 800×g for 10 min. Each sample was re-suspended in a lysis buffer (8 M urea, 4% CHAPS, 1% IPG buffer, 65 mM dithiothreitol [DTT]) containing trypsin (VA9000, Promega, Madison, WI, USA) and a protease inhibitor cocktail (Roche, Basel, Switzerland) with a tiny quantity of silica beads, followed by ultrasonication (UP50H, Hielscher Ultrasonics GmbH, Teltow, Germany) under 40 Hz, 60 pulses for 15 min on ice. Thereafter, the extracted crude proteins were further purified using ReadyPrep 2-D Cleanup kit (Bio-Rad Laboratories, Hercules, CA, USA) and underwent a reductive alkylation reaction. The quantification of the protein extracts was measured by the Bradford method [21].
Proteins extracted from fresh and frozen-thawed semen (1 × 109 sperm cells/ml of each sample) were incubated for 20 min in 2 M thiourea, 500 µL of 8 M urea, 65 mM DTT, 2% CHAPS and protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). After denaturation, proteins (30 μg) were loaded into the top of 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). The molecular weight standard ladder was obtained from Thermo Scientific (PageRulerTM Plus Prestained Protein Ladder, 10 to 250 kDa).
Cation-exchange ProteinChip CM10 Spin Columns (Bio-Rad Laboratories) were used to separate fresh and frozen-thawed sperm surface proteins. Semen samples with 300 µL of binding buffer were left at an ambient temperature for 30 min, centrifuged at 3,200×g for 30 s to remove the reaction frame, and washed three times with 500 µL of binding buffer. Total proteins were collected by eluting with NaCl solutions at concentrations of 0.5, 1, and 2 M.
A portion of the protein combination (100 µg) was once again precipitated in cold acetone before being dissolved in 50% TEAB (Sigma-Aldrich, St. Louis, MO, USA) with 1% SDS. Using The bicinchoninic acid protein test, the samples’ protein content was measured, and the samples were kept in a freezer at −80°C for iTRAQ analysis. The 8-Plex iTRAQ labeling kit’s procedure was followed when the protein pellets were dissolved in 30 L of 50% TEAB with 70 l of isopropanol and tagged (FS360-20-10-CE; Applied Biosystems, Waltham, MA, USA). During the labeling, the digested protein samples of fresh and frozen-thawed semen were given the iTRAQ tags 117 and 119, respectively. Pooling and drying were then treated on iTRAQ-labeled samples in a vacuum freeze-dryer for surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS).
ProteinChip array (Ciphergen Biosystems, Fremont, CA, USA) was used for fresh and frozen-thawed semen samples, with cation exchange (CM10), anion exchange (Q10), and hydrogen surface (H50) arrays. The binding buffer used was 100 mM NaCl (pH 4.0) for CM10, 100 mM Tris-HCl (pH 9.0) for Q10, and 0.5% trifluoroacetic acid in 10% acetonitrile for H50. After air-drying with removal of the biosensor, the sample spot was sprayed with 1 μl of 50% sinapinic acid and analyzed using a PBSIIc Protein Chip Reader (Serise-4000, Ciphergen Biosystems, Fremont, CA, USA). Finally, the output data were recorded at about 150–200 laser wavelengths per section. For molecular mass detection, accuracy of PBSIIc, precise calibration was performed using the ProteinChip Protein Calibrant Kit (Cat No., C10-00001, Bio-Rad Laboratories). To reduce the fluidity of the result, the experiment was conducted simultaneously, and samples under the same conditions were pulled out. Triplicate profiles per sample were obtained by SELDI-TOF MS.
The sperm proteins that were expressed differentially between fresh and frozen-thawed semen were manually annotated using two functional annotation tools, UniProtKB and DAVID. Gene Ontology (GO) terms and pathways for identified proteins were retrieved using the downloaded UniProtKB database, which contains a total of 172,062,311 entries. These proteins’ functional analysis was done using the Database for Annotation, Visualization, and Integrated Discovery (DAVID Bioinformatics Resources 6.8). We also conducted GO enrichment and network analyses using ClueGO (v. 2.5.8) in Cytoscape v. 3.9.1. Enrichment of GO terms was assessed by a two-sided hypergeometric test corrected using the Bonferroni step-down approach and GO terms with a corrected p < 0.05 were considered significant. Additionally, we used a Kappa score (term/pathway connectivity) of 0.4 in advanced selection options. To elucidate the interactions among differentially expressed sperm proteins, an interaction network was designed using STRING online database (v11.5).
RESULTS
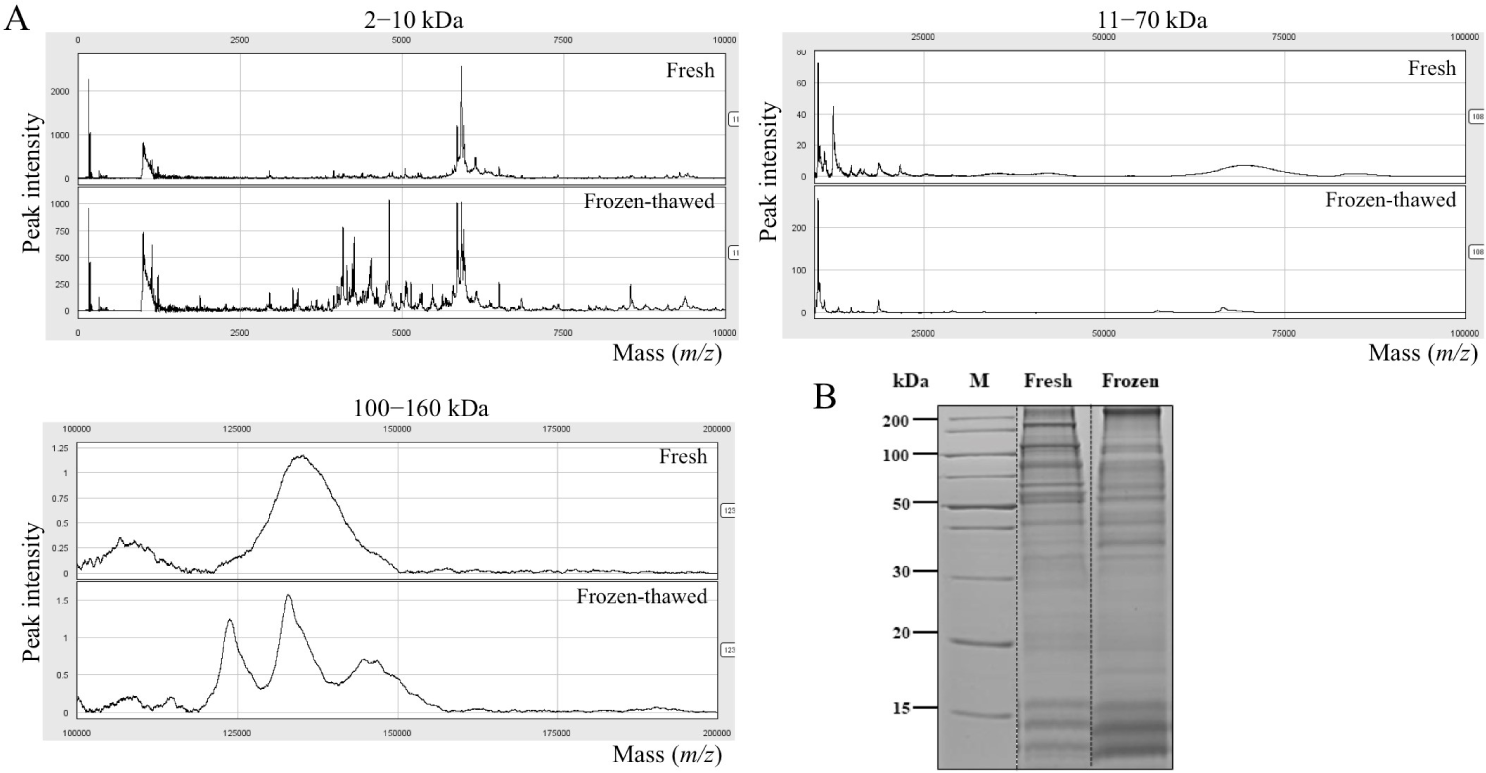
SELDI-TOF MS confirmed the proteins that differed in expression between fresh and frozen-thawed boar semen. Approximately 150 protein peaks were found for each of the CM10, Q19, and H50array. The CM10 array showed more differentially expressed protein peaks (>3-fold change) between the fresh and frozen-thawed semen, among the proteinChips above mentioned (Fig. 1A). Purification and elution were carried out with fresh and frozen sperm through into the same cation exchange proteinChip CM10 spin column. The overall separation patterns of the differentially expressed proteins were observed by SDS-PAGE (Fig. 1B). The efficiency of labeling with iTRAQ reagents was 93.9%, and 76 proteins were identified to be expressed differentially between fresh and frozen-thawed semen using iTRAQ. The entry names were obtained from UniProtKB for all of the proteins, and Table 1 provides a list of the names and functions of these proteins. Nine proteins were more strongly expressed in frozen-thawed samples than in fresh ones, while the opposite pattern was found for 67 proteins.
All GO terms associated with biological processes (a), cellular components (b), and molecular functions (c) relating to the 76 proteins differentially expressed between fresh and frozen-thawed boar semen were generated using DAVID. The eight main GO terms in the category of biological processes included the regulation of nitric oxide biosynthetic process (1), glycolytic process (2), response to unfolded proteins (3), protein kinase A signaling (4), sperm capacitation (5), respiratory electron transport chain (6), catalytic activity (7), and mitochondrial electron transport (8) from NADH to ubiquinone. The cellular component ontology category describes the location and levels of subcellular structures, and macromolecular complexes of proteins. The identified 76 proteins that were differentially expressed between fresh and frozen-thawed semen were localized mostly in the acrosomal membrane, sperm principal piece, mitochondrial matrix, acrosomal vesicle, sperm fibrous sheath, plasma membrane raft, pyruvate dehydrogenase complex, and germinal vesicle. Regarding molecular functions, DAVID analysis also revealed that 3-hydroxyacyl-CoA dehydrogenase activity, cytochrome c oxidase activity, enoyl-CoA oxidase activity, long-chain-3-hydroxacyl-CoA dehydrogenase activity, peptidyl-dipeptidase activity, protein kinase A binding, carboxypeptidase activity, and pyruvate dehydrogenase (NAD+) activity were the most relevant molecular functions of the proteins differentially expressed between fresh and frozen-thawed semen.
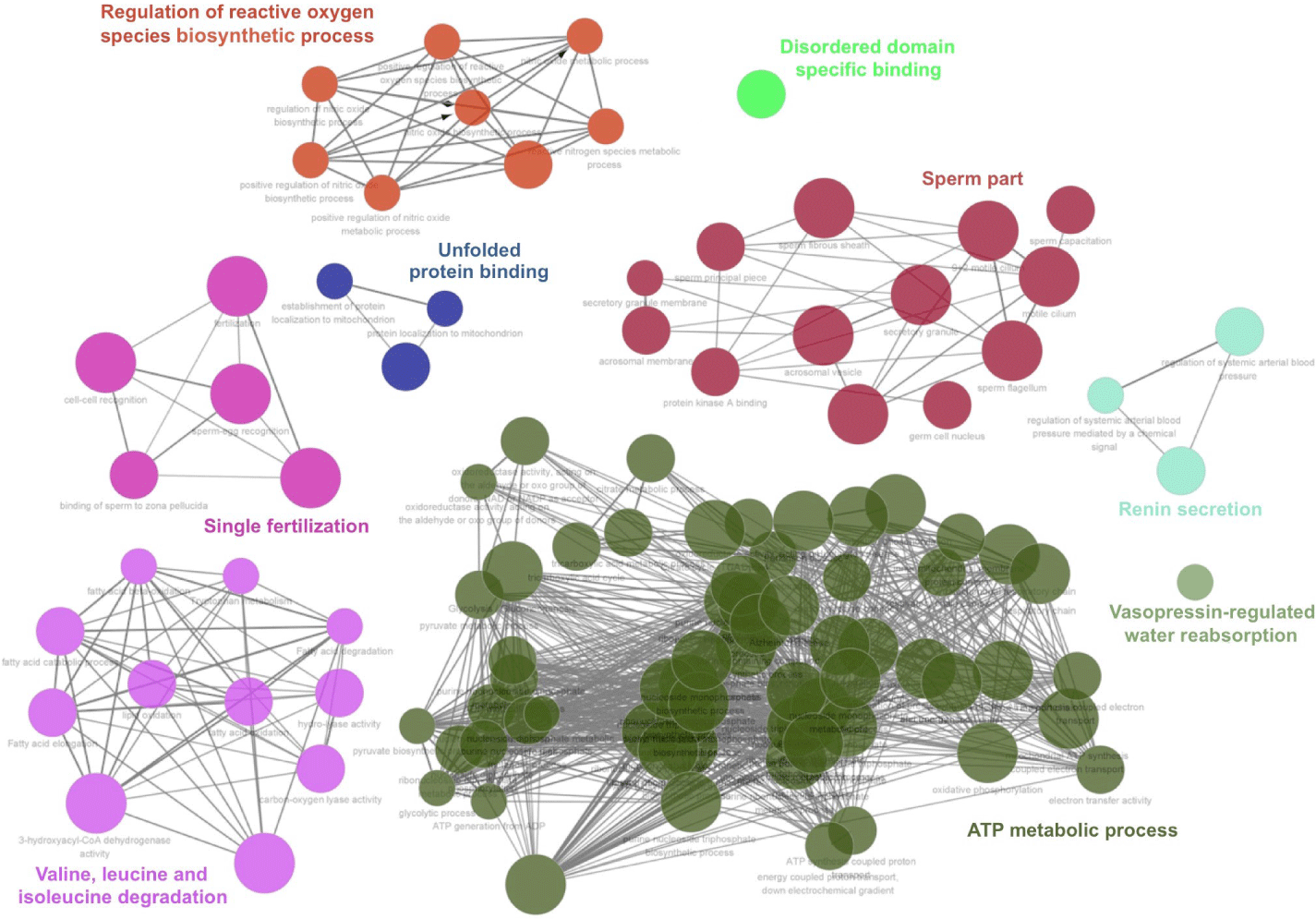
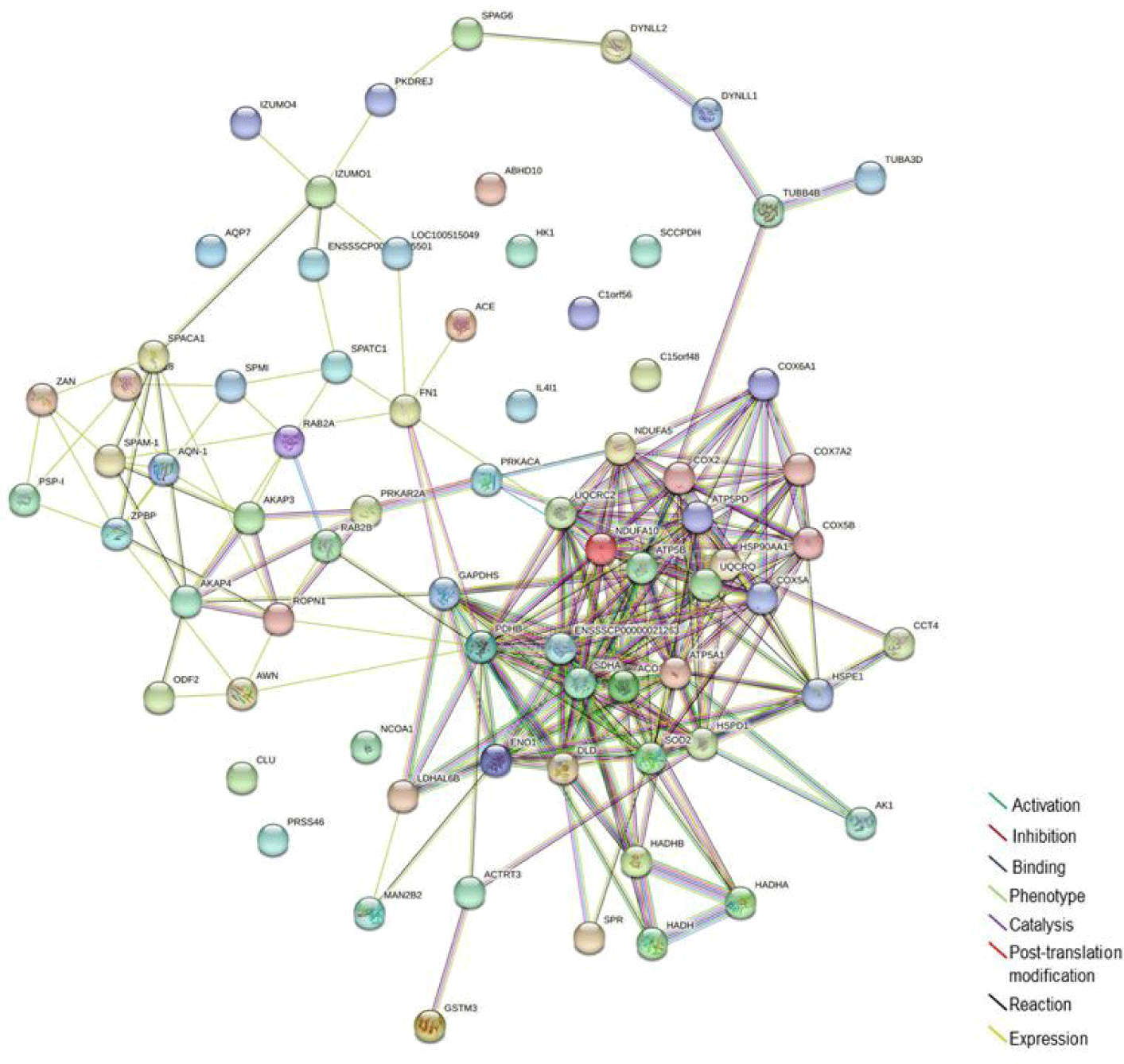
A functional interaction network of the 76 identified proteins was generated using Cytoscape software. The Cytoscape plugin Clue GO allowed analysis of the GO and biological processes in context with other interacting proteins. Of the 76 proteins, 63 were successfully mapped to the UniProtKB database, while this failed for the other 13. Our network map revealed that the differentially expressed proteins mainly participated in oxidative phosphorylation, mitochondrial inner membrane and matrix, pyruvate metabolic processes, sperm flagellum, motile cilium, extracellular organelle, unfolded protein binding, and single fertilization (Fig. 2). In an additional comprehensive analysis of the molecular network among the 76 identified proteins, direct and indirect molecular interactions were evaluated using STRING version 11. On the basis of the STRING online network analysis website, we uploaded the 76 genes to the STRING database, resulting in successful integration of 63 genes into one module (Fig. 3).
DISCUSSION
The conventional approach for exploring the mechanism behind cryo-damage of sperm is 2D gel electrophoresis. To detect quantitative variations in semen proteins after cryopreservation, it is being used in animal species including human [13], sheep [15], chicken [16], carp [17], and sea bass [14]. Considering the expression characteristics of proteins identified through 2D gel electrophoresis, the degree of expression of protein spots is not consistent due to the difference in the analytical conditions and hand rings of the workers [22]. Gel-free approaches are increasingly used for the profiling and quantification of proteins/peptides in a high-quality manner [23]. Furthermore, SELDI-TOF MS has advantages in basic research discovery of protein–protein interactions, As well as suggesting rapid protein expression profiles from various biological samples [23]. A robust proteomic method, such as iTRAQ, can produce analytical data to discover proteins that are produced differently and clarify the biochemical and biophysical processes that underlie animal quality control [18].
To this end, the iTRAQ 2D LC-MS/MS technique was used to perform comparative protein profiling on fresh and frozen boar sperm. We demonstrated more differentially expressed proteins in the CM10 sequence (Fig. 1A). Overall, the fresh sperm showed a high pick. however, a part of the frozen-thawed sperm showed a higher pick than the fresh sperm. Other studies suggested that cryopreservation-induced membrane reorganization and ultrastructural changes in the sperm plasma membrane may lead to bilayer faults that could facilitate an influx of extracellular calcium into the semen [24,25]. It seems the value of some factors has increased as the sperm membrane has adapted to the changing environment during cryopreservation. Also, SDS-PAGE electrophoresis revealed the overall pattern of differentially expressed proteins (Fig. 1B). A total of 76 proteins that were expressed differentially were found and measured, including 9 proteins present at higher levels in fresh sperm (Table 1) and 67 proteins present at higher levels in frozen-dissolved sperm (Table 1). A functional interaction network of these proteins was designed using Cytoscape software. Cytoscape’s CleGO plug-in allowed for finding associated protein networks using a database. The biological process grouped 76 proteins into nine nodes for association, and described the nodes according to their main functions. It was revealed that they mainly participated in oxidative phosphorylation, mitochondrial inner membrane and matrix, pyruvate metabolic processes, sperm flagellum, motile cilium, extracellular organelle, unfolded protein binding, and single fertilization (Fig. 2). The protein-protein interaction network was analyzed by STRING. The analysis established a network according to the structural, functional, and evolutionary properties of proteins and provided a new direction for future experimental studies. After cryopreservation, according to string analysis, the majority of proteins were connected to different sperm features, including sperm energy metabolism, sperm structure, and fertilization. In particular, the classified functions included oxidative phosphorylation, mitochondrial inner membrane and matrix, and pyruvate metabolic processes, which are involved in adenosine triphosphate (ATP) synthesis; and sperm flagellum and motile cilium, which are involved in sperm tail structure. (Fig. 3). Therefore, the proteins differentially expressed between fresh and frozen-thawed boar semen may be involved in sperm energy metabolism and flagellum structure. By exploiting the advantages of iTRAQ, it is possible to quantify multiple protein samples simultaneously in the same LC-MS/MS run.
Chen et al. [19] reported that 41 proteins were identified as being differentially expressed between fresh and frozen-thawed Yorkshire boar semen, of which 35 proteins were expressed more highly in frozen-thawed semen than in their fresh counterparts, while the opposite pattern was shown for 6 proteins. The high/low ratio (35/6; 5.83) of differentially expressed proteins in their study was not dissimilar to our ratio (67/9; 7.44, Table 1). The difference in the total number of proteins identified as being differentially expressed may be due to the difference in fractionation column, LC-MS/MS system, and/or annotation tool, rather than the boar strain. The high sensitivity of boar semen to cryopreservation or cold shock may be associated with the large volume of ejaculate and the fragile nature of the semen membrane [9,10], which is deficient in cholesterol molecules and abundant in unsaturated phospholipids.
In conclusion, our results in this study suggest possible network formation of biomarkers associated with survival rates after freeze-thawing of boar semen, and are expected to provide more accurate information on expression proteins in the distribution of differential proteins expressed after freeze-thawing of boar sperm and to contribute to research related to sperm survival after freeze-thawing.
















