RESEARCH ARTICLE
Complete genome sequence of Pediococcus acidilactici CACC 537 isolated from canine
Jung-Ae Kim1,2
,
Hyun-Jun Jang1
,
Dae-Hyuk Kim1,3
,
Youn Kyoung Son4
,
Yangseon Kim1,*
Author Information & Copyright ▼
1Department of Research and Development, Center for Industrialization of Agricultural and Livestock Microorganisms, Jeongeup 56212, Korea
2Department of Bioactive Material Science, Jeonbuk National University, Jeonju 54896, Korea
3Department of Molecular Biology, Department of Bioactive Material Science, Institute for Molecular Biology and Genetics, Jeonbuk National University, Jeonju 54896, Korea
4Biologicla and Genetic Reources Assessment Division, National Institute of Biological Resources, Incheon 22689, Korea
*Corresponding author: Yangseon Kim, Department of Research and Development, Center for Industrialization of Agricultural and Livestock Microorganisms, Jeongeup 56212, Korea. Tel: +82-63-536-6712, E-mail:
yangseon@cialm.or.kr
© Copyright 2023 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Oct 17, 2022; Revised: Nov 03, 2022; Accepted: Nov 12, 2022
Published Online: Sep 30, 2023
Abstract
Pedi coccus acidilactici CACC 537 was isolated from canine feces and reported to have probiotic properties. We aimed to characterize the potential probiotic properties of this strain by functional genomic analysis. Complete genome sequencing of P. acidilactici CACC 537 was performed using a PacBio RSII and Illumina platform, and contained one circular chromosome (2.0 Mb) with a 42% G + C content. The sequences were annotation revealed 1,897 protein-coding sequences, 15 rRNAs, and 56 tRNAs. It was determined that P. acidilactici CACC 537 genome carries genes known to be involved in the immune system, defense mechanisms, restriction-modification (R-M), and the CRISPR system. CACC 537 was shown to be beneficial in preventing pathogen infection during the fermentation process, help host immunity, and maintain intestinal health. These results provide for a comprehensive understanding of P. acidilactici and the development of industrial probiotic feed additives that can help improve host immunity and intestinal health.
Keywords: Pediococcus acidilactici; Canine; Whole-genome sequencing
Lactic acid bacteria (LAB) are mostly used as probiotics in functional foods and feed additives [1]. Among them, Pediococcus sp. is used as a beneficial microorganism in the context of food and livestock microbiology, and it has been reported that P. acidilactici CACC 537 (KACC 8198BP) has acid and bile tolerance, intestinal adhesion activity, and antibacterial activity against livestock pathogens [2].
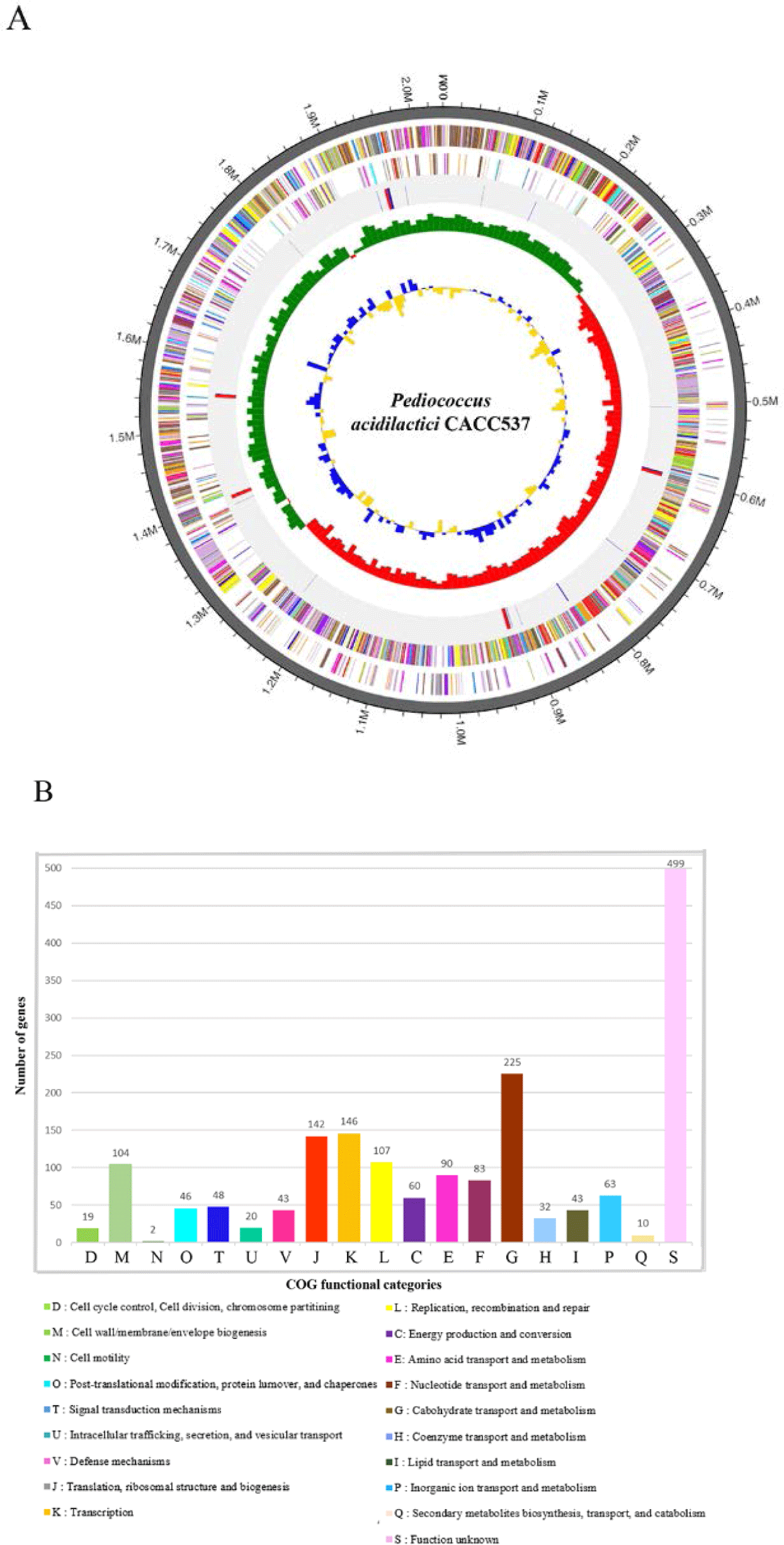
This study attempted to genetically determine the useful effects and functions of CACC537 using whole-genome sequencing. The complete genome of CACC 537 was sequenced using a PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) and Illumina (San Diego, CA, USA) platform with a SMRTbellTM template at Macrogen (Seoul, Korea), and the reads were assembled of the using HGAP version 3.0 [3]. The annotation of the sequence used automatic results from the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) and analyzing Rapid Annotations using Subsystems Technology (RAST) tools [4]. The complete genome of CACC537 consists of one circular chromosome with a total length of 2,035,984 bp and a DNA G + C content of 42.0%. it includes 1,897 protein-coding genes (CDSs) and 71 RNA genes (15 rRNA and 56 tRNA genes) (Table 1 and Fig. 1A). The 1,782 genes were categorized as functional proteins based on designations through the Clusters of Orthologous Groups (COGs) database. Most assigned CDSs were found to be involved in carbohydrate transport and metabolism; transcription; translation, ribosomal structure and biogenesis; replication, recombination and repair; and cell wall/membrane/envelope biogenesis (225, 146, 142, 107, and 104 genes, respectively) (Fig. 1B).
Table 1.
Genome overview of Pediococcus acidilactici CACC 537
| Feature |
Values |
| Genome size (bp) |
2,035,984 |
| No. of contigs |
1 |
| GC content |
42.0 % |
| Protein-coding genes (CDSs) |
1,897 |
| rRNA |
15 |
| tRNA |
56 |
| Genbank Acession No. |
CP048019 |
Download Excel Table
Fig. 1.
Genome features of Pediococcus acidilactici CACC 537.
(A) Circular genome maps of the P. acidilactici CACC537 chromosome. Circles from the outside to the center denote rRNA and tRNA genes, reverse strand CDSs, forward strand CDSs, GC skew, and GC content. (B) COG functional category annotation numbers on CACC 537 genome. CDS, coding sequences; GC, guanine cytosine; COG, clusters of orthologous groups.
Download Original Figure
In the genome of strain CACC 537, the presence of ldhD, ldhL, pyk, eno, fbaA, pfk, and pgi, which are involved in lactic acid production as key genes of homo-fermentation in the Embden-Meyerhof-Parnas (EMP) pathway, was detected [5]. We also identified genes encoding lysozymes, chitinases, and proteases, commonly predicted as able to exert antibacterial functions against pathogens; and the fab gene cluster, in charge of producing fatty acids that can protect the intestinal mucosal from pathogens. Interestingly, the short-chain fatty acids (SCFA) produced as metabolic by-products play an important role in intestinal homeostasis and have been reported to be associated with immune enhancement and liver function improvement [6–8]. In addition, we identified a type II-A CRISPR/CRISPR-related (Cas) gene with two CRISPR (1 and 2) regions, and a CRISPR 9 region; and genes involved in immune and defense mechanisms, including some for restriction-modification (R-M) systems. This strain prevents fermentation failure due to bacterial phage and pathogen infection in the fermentation process, and has an intestinal function to help host immunity and maintain intestinal health [9–10] (Table 2).
Table 2.
Predicted genes involved in probiotic potency in Pediococcus acidilactici CACC537
| Predicted function |
Pediococcus acidilactici CACC 537 |
| Predicted genes |
Products |
Gene position |
Length (bp) |
| Homolactic-fermentation related |
|
|
ldhD
|
D-Lactate dehydrolase |
195437..196432 |
996 |
|
ldhL
|
L-Lactate dehydrolase |
940686..941657 |
972 |
| c(1521565..1522485) |
921 |
|
pyk
|
Pyruvate kinase |
c(408957..410720) |
1,764 |
|
eno
|
Phosphopyruvate hydratase |
c(1111207..1112493) |
1,287 |
| 1802875..1804197 |
1,323 |
|
fbaA
|
Fructose-bisphosphate aldolase |
730570..731433 |
864 |
|
pfk
|
6-Phosphofructokinase |
c(410797..411765) |
969 |
| c(552661..553572) |
912 |
|
pgi
|
Glucose-6-phosphate isomerase |
c(727794..729146) |
1,353 |
| Antimicrobial resistance-related |
| Bacteriolytic enzyme |
|
|
- |
Lysozyme |
c(465359..466543) |
1,185 |
| - |
Chitinase |
1707522..1708034 |
513 |
| 2026125..2027144 |
1,020 |
|
glup
|
Rhomboid protease |
125912..126601 |
690 |
| Fatty acid biosynthesis (Clusters) |
|
|
fabI
|
Enoyl-(acyl-carrier-protein) reductase |
187972..188730 |
759 |
|
accA,D,C
|
Acetyl-CoA carboxylase |
184990..187955 |
2,966 |
|
fabZ1
|
3-Hydroxyacyl-(acyl-carrier-protein) dehydratase |
184563..184973 |
411 |
|
accB
|
Acetyl-CoA carboxylase |
184125..184550 |
426 |
|
fabF,G,D
|
β-Ketoacyl-ACP synthase II, β-ketoacyl-ACP reductase, acyl-carrier protein |
181121..184120 |
3,000 |
|
acpP
|
Acyl carrier protein |
180858..181094 |
237 |
|
fabH,Z2
|
β-Ketoacyl-ACP synthase III, β-hydroxyacyl-ACP dehydratase |
179410..180831 |
1,422 |
| Restriction-modification system |
|
|
hsdR
|
Type I site-specific deoxyribonuclease |
1445338..1448211 |
2,874 |
|
hsdM
|
Site-specific DNA-methyltransferase |
1448232..1449824 |
1,593 |
|
hsdS
|
Type I site-specific deoxyribonuclease |
1449821..1453105 |
3,284 |
| Immune system |
|
|
cas9
|
CRISPR-associated endonuclease |
1747125..1751225 |
4,101 |
|
cas1
|
CRISPR-associated endonuclease |
1751443..1752348 |
906 |
|
cas2
|
CRISPR-associated endoribonuclease |
1752326..1752631 |
306 |
|
yajC
|
Immunogenic membrane protein |
c(647443..647826) |
384 |
|
cfa
|
Cyclopropane-fatty-acyl-phospholipid synthase |
1113749..1114924 |
1,176 |
|
dacA
|
Diadenylate cyclase |
1814096..1814947 |
852 |
|
myh6
|
TNF receptor protein |
1730618..1733290 |
2,673 |
Download Excel Table
This study on the complete genome sequence of P. acidilactici CACC 537 showed that it has potential probiotic effects, and thus may be useful for the development of health-promoting products.
Nucleotide sequence accession number
The complete genome of P. acidilactici CACC 537 has been deposited in the NCBI GenBank database under the accession number CP048019.