INTRODUCTION
Antibiotic resistance (AR) is a complex, multifaceted issue that poses a serious threat to the health of livestock and humans, as well as the surrounding environments, including soil, air, and water. Its intricacy results from various interrelated elements that contribute to its development and spread, making it a challenging issue to address. AR is on the rise globally, and if current trends continue, there could be up to 10 million annual AR-associated deaths from a variety of untreatable infections by 2050 [1,2]. It is one of the many concerns of the 21st century identified by the Lancet Commission that threatens to hinder future progress in global health [3].
AR is one of the significant global health issues that best exemplifies the One Health concept [4]. This is because bacterial species such as Escherichia coli (E. coli), Salmonella and Campylobacter carry antibiotics resistant genes and can spread between livestock, humans, and the environment [5-7]. The One Health approach recognize the interconnection and mutual influence of these three domains. One Health is defined as “a collaborative effort of multiple-discipline health science professions, together with their allied disciplines and institutions-working locally, nationally, and worldwide to achieve optimal health for domestic animals, humans, wildlife, plants, and the environment as a whole” [7]. AR is closely linked to each of these factors due to the reckless and excessive use of available antibiotics in agriculture, livestock farming and human medicine [8,9]. The interconnections between livestock, humans, and environmental compartments create pathways for the transmission of bacteria, mobile genetic elements (MGEs), and antibiotics themselves [10]. These pathways facilitate the movement of these entities between these compartments, thereby promoting the spread of AR.
The local and global spread of AR can be attributed to various factors, including inadequate antibiotic use in both livestock and humans, contaminated environments, and ineffective infection control policies [11-13]. Careless human use of antibiotics contributes to the development and spread of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs). These resistant bacteria can be transmitted to other humans through the direct contact, as well as through food, water, and environmental surfaces [14]. Similarly, the use of antibiotics in livestock can also contribute to the emergence of antibiotic-resistant bacteria. These bacteria can be transmitted to humans through direct contact with livestock, via food products, and can also be present in the environment due to livestock waste [15]. However, it is not yet clear to what extent livestock-derived transmission routes contribute to the spread of AR compared to human-derived routes [10]. Environmental contamination is also a major factor in the dissemination of antibiotic-resistant bacteria and genes. Antibiotic-resistant bacteria can persist in soil and water, potentially spreading to animals and humans when they come into contact with contaminated environmental sources [16]. While selective pressure and horizontal gene transfer are known factors in the spread and dissemination of antibiotic-resistant bacteria and genes, the absence of proper policies and regulations for the use and disposal of antibiotics only exacerbates this issue.
This review aims to enhance understanding of the interdependence between livestock, human, and environmental health in terms of the emergence, spread, and concerns related to AR in each of these domains.
ANTIBIOTIC RESISTANCE IN LIVESTOCK
ARB and ARGs in food animals are regarded as emerging contaminants that pose a serious threat to global public health [17]. Multiple reports have indicated a 35% increase in global antibiotic consumption during the first decade of the 21st century [18]. A study conducted by Muchandani et al. using statistical models estimated antimicrobial usage at 99,502 tons in 2020 and projected an 8% increase to 107,472 tons based on the latest trend [19].
Livestock farms indeed represent a significant source of ARB and ARGs. These ARBs often display resistance to several major classes of antibiotics, such as aminoglycosides, tetracyclines, sulfonamides, chloramphenicols, and β-lactams [20,21]. Livestock farms can serve as reservoirs for pathogenic zoonotic bacteria such as Staphylococcus aureus (S. aureus), Salmonella species, Campylobacter species, E. coli, andListeria species. These pathogens can be transmitted into environment through air, slurry, and feces from livestock, leading to the potential outbreak of infections and acting as a source of ARGs [22,23].
The causes of AR are indeed very complex, involving various factors such as the overuse and misuse of antibiotics in both human medicine and agriculture, as well as environmental contamination [24]. The administration of antibiotics in livestock is primarily motivated by three main factors: to treat infections, to prevent animals at risk from getting infection or as feed additives to boost growth [25]. Numerous studies have demonstrated a link between the use of antibiotics in food-producing animals, especially for growth promotion, and AR in people who either live on or near farms [15]. Additionally, drug-resistant strains can emerge if the prescribed antibiotics are not changed in a timely manner [26]. These bacteria generally develop resistance to antibiotics through several different resistance mechanisms, including blocking the antibiotic’s target sites, altering the permeability of cell membranes, and producing antibiotic-inactivating enzymes [27]. The resistance observed in food-related bacteria reflects the broader condition of bacterial resistance within the environment from which food for human consumption originates [28]. Despite an increase in the number of relevant studies on livestock and AR, as of 2020, they still accounts for only 17% of all the studies published on AR [29]. Therefore, there is a pressing need to promote more research in this area to better understand and address the complex issues surrounding AR in livestock and its implications for human health.
ANTIBIOTIC RESISTANCE IN HUMANS
The human community is facing an escalating crisis of antibiotic-resistant infections in patients, presenting a persistent and increasing threat to public health [30].
The World Health Organization (WHO) released a list of 12 families of bacteria in 2017 that pose a significant threat to human health [31], categorizing them into three priority groups based on the urgency of developing novel antibiotics to combat them [32]. The most critical group comprises multidrug-resistant bacteria such as Acinetobacter, Pseudomonas, and certain Enterobacteriaceae like Klebsiella pneumoniae (K. pneumoniae), E. coli, and Enterobacter species. These bacteria can cause serious and potentially fatal infections, especially in patients admitted to hospitals and nursing homes or those requiring ventilators and catheters, leading to conditions such as pneumonia and bloodstream infections [33-35]. The high-priority category, which includes bacteria such as Enterococcus faecium (E. faecium) and S. aureus, exhibits resistance to a number of antibiotics such as vancomycin and fluoroquinolones [33]. The medium-priority category includes Streptococcus pneumoniae and Shigella, which may have some resistance but can still be effectively treated with the antibiotics currently available [36]. Table 1 lists the priority pathogens identified by the WHO for the development of new antibiotics to combat antibiotic-resistant bacteria (Table 1).
AR is indeed a natural process, but its uncontrolled spread has escalated into a public health emergency largely due to the overuse and misuse of antibiotics. This excessive usage accelerates the development and dissemination of antibiotic-resistant bacteria [37]. Among the most influential human behaviors contributing to the development and spread of AR are the failure of patients to adhere to prescribed antibiotic regimens and their tendency to engage in self-medication. These behaviors have a significant impact on the proliferation of antibiotic-resistant strains [38]. The public’s perception that antibiotics offer a quick and effective solution to common health problems has led to actions that undermine a doctor’s prescribing control [39]. The use of broad-spectrum antibiotics as a substitute for accurate diagnostics or to increase the chances of therapeutic success also contributes to the selection of resistant bacteria [40]. According to a report from the Centers for Disease Control and Prevention (CDC), more than 50% of antibiotic prescriptions are considered unnecessary in the United States [41]. While the increase in the prevalence of AR cannot be solely attributed to antibiotic overuse, other factors known as “socioeconomic determinants” also play a significant role. These determinants include poor community hygiene, unsafe food practices, inadequate infection control measures in healthcare settings, the presence of antibiotics in the environment, and their use in animals and agriculture [42]. AR in humans is also influenced by entrenched medical practices, economic pressures, and vaccine hesitancy [43].
ANTIBIOTIC RESISTANCE IN ENVIRONMENT
In recent years, AR in the environment has emerged as a significant public health concern [44]. Several studies have reported the presence of ARB and antibiotic residues in various environmental samples, including soil, water, and wildlife. These findings underscore the need for effective strategies to mitigate the further spread of AR in the environment [44,45]. AR can arise through mutations in a bacterium’s pre-existing genome or through the uptake of foreign DNA, both of which contribute to the development of AR. Mutations can easily occur in bacteria in a livestock or patient receiving antibiotic treatment and can become permanently established. However, such intense selection pressure on pathogens is less common in other environmental settings. Therefore, external environmental factors are less likely to influence mutation-based evolution of resistance in most pathogens. In contrast, the uptake of novel resistance factors is facilitated by the diverse ecological niches of water, soil, and other environments, creating a broad gene pool that surpasses the microbiota of livestock and humans [46,47].
The detection of ARGs in diverse environmental settings, including sludge, wastewater, river water and sediment, air, and soil has been well documented [48]. Soil, in particular, serves as a natural habitat for a diverse range of microorganisms and is considered the primary and natural reservoir of ARGs. This presence of ARGs in bacteria without prior exposure to antibiotics is commonly referred to as intrinsic resistance. It highlights the fact that these resistance genes can be naturally occurring and not solely the result of antibiotic use or pollution [49]. However, due to human activities, soil has transformed from being primarily an intrinsic reservoir of ARGs to also serving as a repository and accumulation site for numerous acquired ARGs [41]. A comparative study of soil microbiome in natural and agroecosystems revealed that natural ecosystems had a relatively lower presence of ARB. The study also revealed that Pantoea agglomerans and Bacillus cereus showed the highest levels of resistance. Approximately 23% of the isolated bacteria exhibited resistance to antibiotics [50]. Among all classes of ARGs found in wastewater samples worldwide, tet genes exhibited the highest detection frequency, with researchers identifying approximately 20 different types of tet genes in wastewater [51]. Untreated sewage has also been found to have a variety of ARGs encoding aminoglycosides, β-lactams, tetracyclines, and vancomycin [52-55]. Water has also been affected by ARG contamination. Several types of ARGs have been identified in microbial populations of natural water sources [56,57]. Das et al. discovered the presence of 139 ARGs belonging to 11 different ARG types in the Yamuna riverine ecosystem [58]. A study conducted by Pruden et al. clearly demonstrated the presence of ARGs in numerous environmental compartments in Northern Colorado, including river sediments, irrigation ditch water, dairy lagoon water and even in drinking water treatment plants [44]. Other studies have also identified many different ARBs in environmental samples, including E. coli, K. pneumoniae, and Pseudomonas aeruginosa (P. aeruginosa) [51,59]. Beyond these commonly found bacteria, a study in China isolated New Delhi metallo-β-lactamase 1 (NDM-1) producing Acinetobacter baumannii (A. baumannii) in hospital sewage [60]. Even the air has not escaped AR contamination, as a global survey of ARGs conducted by Li et al. revealed that urban air also contains ARGs, raising various health concerns in cities [61].
Antibiotics make their way into the environment through various pathways, including municipal sewage, hospital waste, animal husbandry refuse, antibiotic production facilities, agricultural runoff, and other sources. A significant portion of antibiotics is excreted in urine and feces within 8–24 hours after consumption, with excretion rates ranging from 30% to 90% [62]. Antibiotics have a variable half-life, ranging from hours to hundreds of days, and even after degradation, antibiotic residues persist in the environment, contributing to the selection for AR [63,64]. Antibiotics are excreted from the human body in the form of antibiotic metabolites, glucuronic acid, and sulfuric acid conjugates. Studies have shown that approximately 70%–80% of antibiotics in sewage remain in a consistent form [45,65,66]. In addition to household and hospital sewage, pharmaceutical production facilities also contribute to the antibiotic burden in sewage treatment plants, as the effluent from the pharmaceutical industry is often inadequately treated before discharge [67,68]. Animal husbandry waste and agricultural runoff are additional sources of antibiotics in the environment [69]. The extensive use of antibiotics in agriculture results in their detection in agricultural runoff, which eventually mixes with water bodies, further contributing to environmental contamination.
DISSEMINATION OF ANTIBIOTIC RESISTANCE AT LIVESTOCK-HUMAN-ENVIRONMENT INTERFACE
AR is a global issue at the livestock-human-environment interface, providing suitable conditions for the rapid spread and evolution of bacteria. The excessive and irresponsible use of antibiotics in medicine, agriculture, livestock farming, and aquaculture has resulted in a rise in resistant bacteria in these areas.
AR evolves and spreads in a complex parameter space and therefore there is still a lack of concrete explanation regarding the spread of AR and the directions of its spread [5,70]. However, AR bacterial clones, MGEs, and ARGs have been found to be responsible for the dissemination of AR at the human-animal-environment interface [11]. MGEs, classified into 2 groups: the intercellular MGE (e.g., plasmids) and the intracellular MGE (e.g., transposons, integrons) act as vehicles for dissemination of resistance at the human-animal-environment interface [71]. Together, these components have a significant role in promoting horizontal genetic exchange, which helps disseminate resistance genes [72]. It has been found that MGEs may have even a greater role in the transmission of AR genes from livestock to humans than bacterial cells [73,74]. K. pneumonia is regarded as a host of a variety of mobile ARGs and is crucial in the global spread of various extended-spectrum β-lactamases and carbapenemase [75]. MGE is also responsible for resistance in E. faecium, S. aureus, A. baumanni, P. aeruginosa, and Enterobacter species [76]. The epidemiology of AR in context of human and animal interactions is also inherently complex, involving numerous potential transmission routes and vehicles. Among these, the foodborne route of transmission is of serious concern, involving pathogens such as Salmonella enterica, Campylobacter coli/jejuni, Yersinia enterocolitica. Bacteria released into the environment from the feces of livestock and humans can disrupt the normal environmental flora, potentially acting as a reservoir before reintroduction into the livestock-human cycle. Therefore, all three domains—livestock, humans, and the environment—are deeply interconnected, and changes in one domain can have far-reaching effects on the others [77].
CONCERNS FOR LIVESTOCK, HUMAN AND ENVIRONMENTAL HEALTH ASSOCIATED WITH ANTIBIOTIC RESISTANCE
AR has emerged as one of the most pressing threats to livestock, public health, and the environment, posing serious obstacles to the successful prevention and treatment of infectious diseases. The fact that infectious diseases induced by ARB are no longer treatable with antibiotics foreshadows an uncertain future in the context of health care [78]. The reasons behind AR are highly complex and involve human behavior at multiple levels of society, with outcomes that affect everyone worldwide, including livestock, the environment, and humans themselves (Fig. 1) [8].
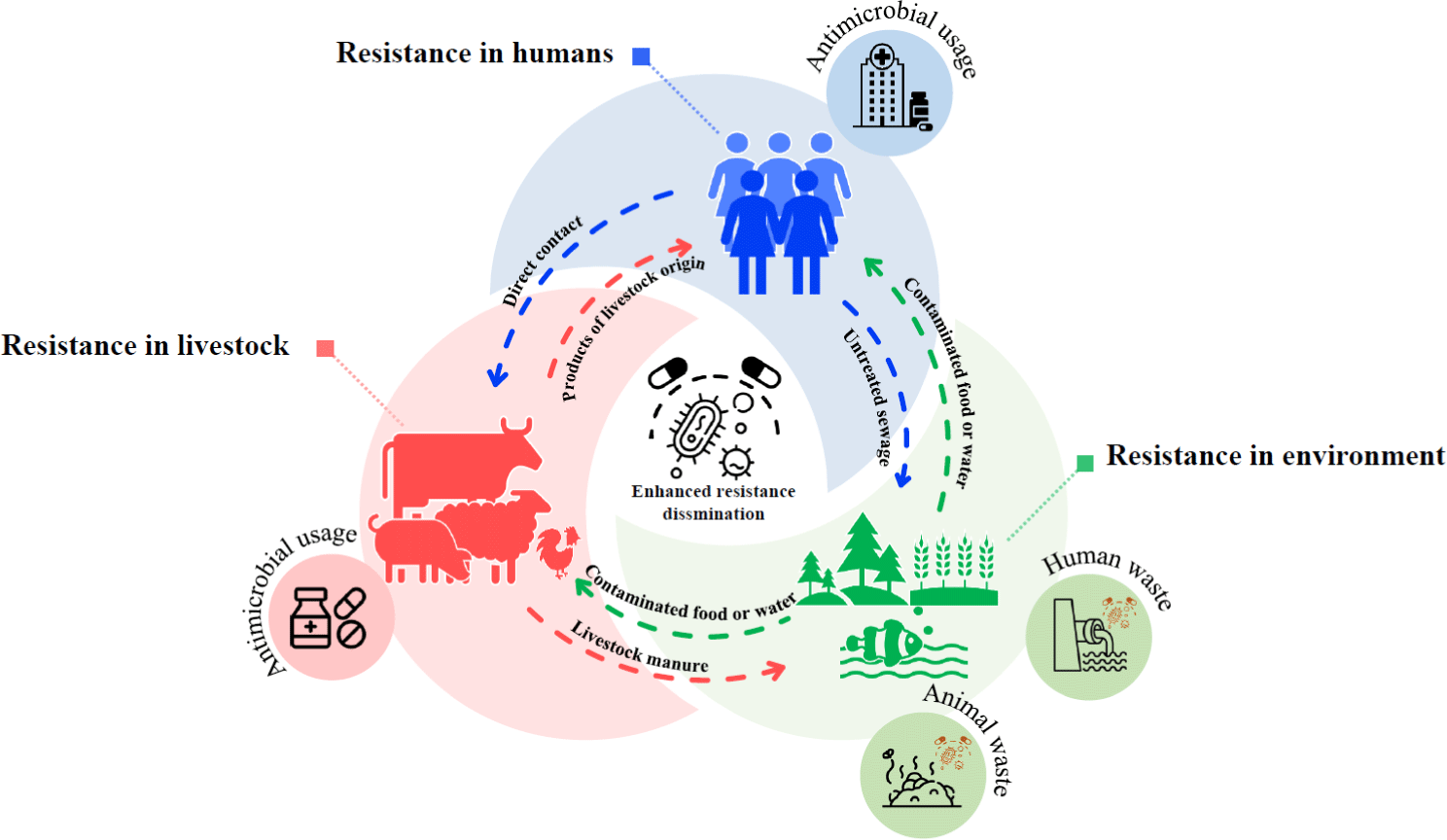
AR has become an ominous specter for humanity. A study in 2019 estimated 1.27 million deaths resulted from resistance in 88 pathogen-drug combinations that were evaluated [79]. Methicillin-Resistant S. aureus (MRSA), a well-known case of AR, has been associated with a high death rate worldwide every year [80]. Additionally, the treatment of various infections, including urinary tract infections and pneumoniae, caused by multi-drug resistant gram-negative bacteria (MDR-GNB) has become more challenging [81,82]. Furthermore, drug resistance to gonorrhea, tuberculosis, and typhoid fever is also on the rise annually, contributing significantly to high healthcare expenses, particularly in developing countries [83].
The worldwide increase in the use of antibiotics as an indispensable part of poultry and livestock production industries, for treating and preventing infectious diseases, as well as for promoting growth, has led to the emergence of AR in bacteria in recent years [84]. This trend significantly impacts the health of both livestock and humans. The extensive use of antibiotics in livestock farming generates antibiotic-resistant bacteria and genes that can potentially transmit to humans through the food chain, posing a threat to the treatment of human infections, particularly fluoroquinolones, which are of major importance in human medicine [85,86]. However, the use of antibiotics from chemical classes different from those used by humans may still be permissible and not contribute to resistance.
The environment serves as a hidden source of antibiotic-resistant organisms carrying resistance genes. This is due to the deposition of antibiotic residues from the pharmaceutical industry, livestock husbandry, hospitals, fecal waste from humans and animals, agricultural runoff, and more. As a result, the environment is responsible for transmitting multi-drug resistant pathogens and ARGs of public health concern [87-90]. Although bacteria exhibited resistance to many drugs before the advent of antibiotics, human activities have exacerbated its spread, leading to the creation of a ‘global resistome’ [91]. Given the large-scale production of antibiotics each year, environmental niches play a significant role in the persistence and spread of AR.
ONE HEALTH STRATEGIES FOR ADDRESSING THE ISSUE OF ANTIBIOTIC RESISTANCE
The One Health paradigm has now become an integral part of global initiatives aimed at confronting AR. Consequently, the major challenges in tackling AR revolve around the differing interests of various social sectors and groups involved in animal, human, and environmental health. These stakeholders have come to a consensus on critical areas of action, the most effective approaches for monitoring AR, containment of infections, and the policies governing the use of antimicrobials [4].
In a collaborative effort to combat AR, the World Organization for Animal Health (OIE), the Food and Agriculture Organization of the United Nations (FAO), and the WHO have introduced the Global Action Plan on AR, facilitating the development of AR-related policies within the One Health framework [92]. These organizations have also established guidelines for AR surveillance to ensure comprehensive monitoring of antimicrobial usage and consumption across animal, human, and environmental settings [93]. Undoubtedly, enhancing global surveillance of drug resistance is essential for effectively combating AR. The medical and scientific community requires a clear understanding of existing and historical AR data to uncover new mechanisms of resistance acquisition, conclusively identify ongoing cases, and anticipate future threats. This task necessitates a deeper understanding of three key aspects: antibiotic consumption in both human and animal contexts, current levels of AR, and the molecular foundations of AR. Additionally, to effectively combat AR through the One Health approach, international collaborations among nations are imperative, as global actions are essential [4].
CONCLUSION
AR is a serious global health concern. To reduce its impact on livestock, human, and environmental health and ensure the longevity of the efficacy of both current and prospective antibiotics, the One Health approach should be emphasized. Recognizing the interrelated nature of these three domains, bringing the human, animal, and environmental sectors together is essential for effectively combating AR and securing the health and development gains achieved over the years.
















