The Enterococci bacteria belong to lactic acid bacteria (LAB) group, which can be found in fermented foods [1]. Especially, Enterococcus faecium is also utilized as probiotics, which could enhance the microbial balance in animals [2]. Despite of safety concerns regarding its use as probiotics, recent research has explored the use of Enterococcus faecium as a feed additive for livestock to enhance growth performance [1,3].
In the present study, the E. faecium strain AK_C_05 was isolated from homemade cheonggukjang, the Korean traditional food, collected from a local market in Cheonan (36.802917° N, 127.149796° E), Chungcheongnam-do, South Korea. Then, the whole genome sequencing was performed to understand the genomic characteristics of E. faecium strain AK_C_05 as a potential probiotic in the livestock industry. The E. faecium strain AK_C_05 was cultivated in Enterococcosel broth (MBcell, Seoul, South Korea) at 37°C for 24 hours. Genomic DNA was extracted from the cultured E. faecium pellet using CTAB DNA extraction method. The complete genome of the E. faecium AK_C_05 was sequenced using the Oxford Nanopore Technologies MinION platform at eGnome (Seoul, South Korea). Briefly, library preparation was performed using Native barcoding Sequencing Kit (SQK_NBD114.24, V14) following the manufacturer’s instructions (Oxford Nanopore Technologies, Oxford, UK). The prepared library was loaded into the MinION MK1b sequencing device (Oxford Nanopore) equipped with a MinION flow cell (MIN114, R10.4.1, Oxford Nanopore). The Oxford Nanopore sequencing produced 79,247 of long reads, resulting in a total of 572,297,864 base pairs. De novo assemble was performed using a Flye assembler v2.9.2, followed by polishing using the Homopolish polisher v0.4.1. The quality of genome assembly was assessed using Quality Assessment Tool for Genome Assemblies (QUAST) v5.2.0 [4]. The quantitative assessment of the genome completeness was conducted using the Benchmarking Universal Single-Copy Orthologs (BUSCO) v5.4.6 [5]. To annotate and predict the protein coding genes, rRNA, and tRNA genes of E. faecium strain AK_C_05, the Rapid Annotation using Subsystem Technology (RAST) v2.0 tool was utilized [6]. The functional categorization of all predicted protein coding genes was performed using the Clusters of Orthologous Groups (COGs)-based EggNOG-mapper v2.0 [7]. Furthermore, the presence of virulence factors and antibiotic resistance in E. faecium strain AK_C_05 was predicted using the BLASTn method, with reference to the Virulence Factor Database (VFDB) and the Comprehensive Antibiotic Resistance Database (CARD) [8,9].
The complete genome of the E. faecium strain AK_C_05 contain one circular chromosome (2,691,319 bp) with a guanine + cytosine (GC) content of 38.3% and one circular plasmid (177,732 bp) with a GC content of 35.48%. A total of 2,827 predicted protein-coding sequence, 18 rRNA genes, and 68 tRNA genes were identified in E. faecium strain AK_C_05. The most abundant COGs category, excluding Function unknown [S], was Carbohydrate transport and metabolism [G], which accounted for 235 genes, representing 10.4% of the total genes identified. The genome feature and map of E. faecium strain AK_C_05 were presented in Table 1, Figs. 1A and 1B.
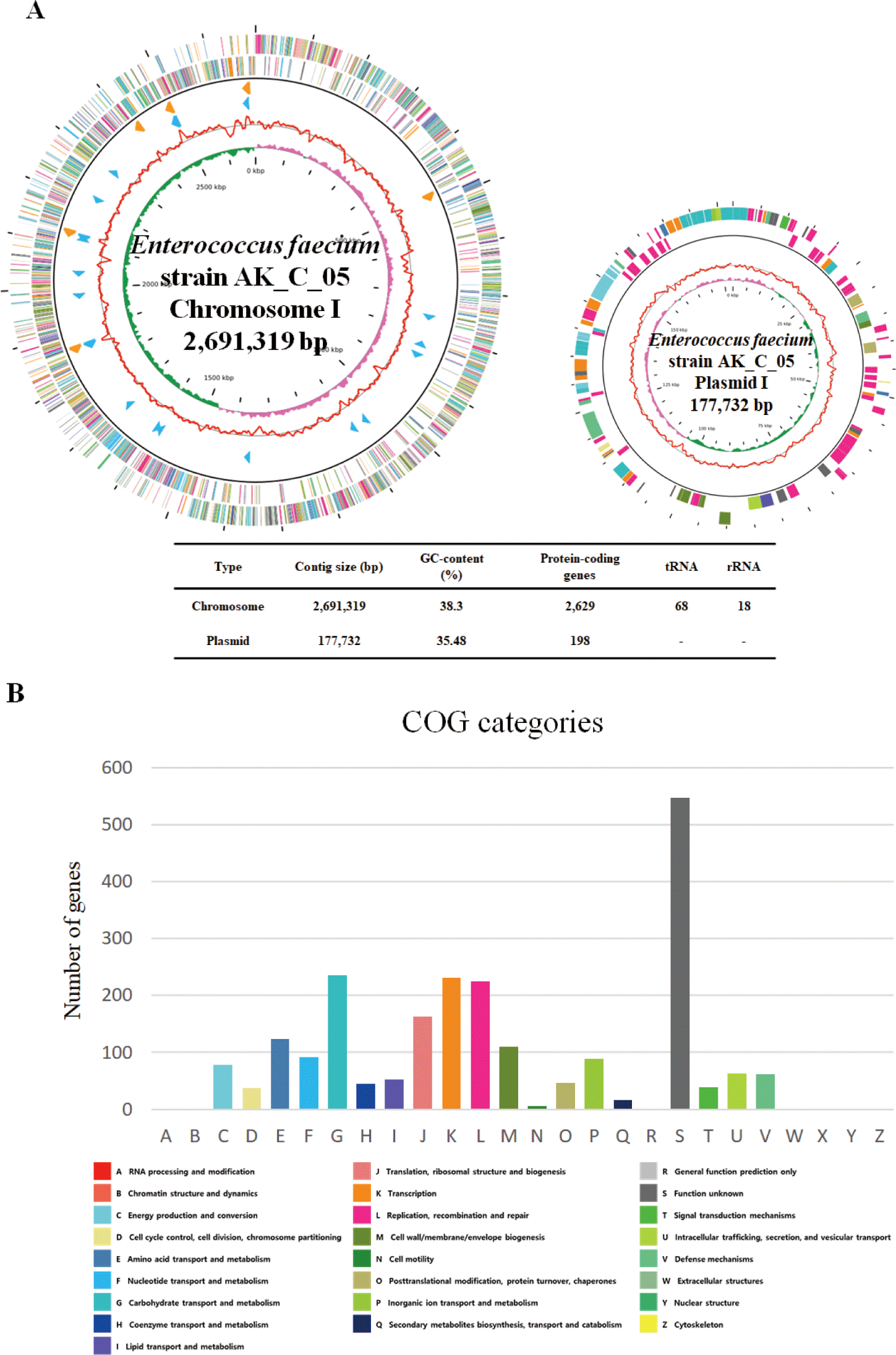
Based on its specific focus on carbohydrate transport and metabolism, E. faecium strain AK_C_05 possesses genes and enzymes, such as alpha-galactosidase (EC 3.2.1.22), beta-glucosidase (EC 3.2.1.21) and alpha-L-arabinofuranosidase (EC 3.2.1.55), that enable efficient utilization of carbohydrates and the capacity to derive energy from diverse carbohydrate substrates. This characteristic makes E. faecium strain AK_C_05 a potential candidate for application in the livestock industry. The complete genome of E. faecium strain AK_C_05 has indicated the presence of the antibiotic resistance gene aac (6’)-Ii in the chromosome and not in the plasmid, confirming that there is no potential for transmission of the resistance gene to other microorganisms. In the plasmid of E. faecium strain AK_C_05, the filA gene was detected, while no other virulence factors were identified. Interestingly, the filA gene’s ability to facilitate adhesion to the cell wall is regarded as a beneficial trait for probiotics [10]. Overall, our results indicate that E. faecium AK_C_05 could be a promising functional probiotic for improving growth performance in the livestock industry.
















