INTRODUCTION
Wheat is one of the most important cereal crops, with a global production comparable only to maize and rice [1]. This in addition to its value as an energy source [2] has made wheat a common ingredient in poultry rations. Although largely composed of starch, the wheat grain contains approximately 10% protein [3,4] and of this, up to 80% is contributed by gluten proteins [5,6].
Wheat gluten proteins are a complex mixture of hundreds of proteins that can be grouped into two approximately equal fractions; gliadins and glutenins [7]. These proteins are also members of the prolamins superfamily of plant proteins which is characterized by a high content of proline and glutamine amino acid residues [7]. When hydrated, gluten proteins confer the rheological properties that make dough good for making bread, biscuits, pasta and as a binder in many processed foods. The viscosity and extensibility of the dough is contributed mostly by the gliadins, while its strength and elasticity come largely from the glutenins [7,8].
Although widely used in human food products, gluten is potentially toxic to some genetically predisposed individuals causing conditions collectively referred to as gluten intolerance. These conditions include; autoimmune celiac disease [9], wheat allergy and non-celiac gluten sensitivity [10]. The high content of proline and glutamine renders the prolamins in gluten partially resistant to hydrolytic action of pancreatic and intestinal proteases [11]. This results in the accumulation of partially hydrolyzed long polypeptide chains that are highly immunostimulatory. The gliadins are also known to induce intestinal permeability through upregulation of zonulin, which breaks down the intestinal epithelial tight junctions [12]. As a result, the peptides reach the lamina propria where their glutamine residues are selectively converted into glutamic acid in a deamidation reaction catalyzed by tissue transglutaminase 2 (tTG2) [13]. The deamidated peptides have an increased negative charge that promotes binding to the specific human leucocyte antigen (HLA-DQ2 or HLA-DQ8) [14]. These peptide-DQ complexes are even more immunostimulatory thus leading to inflammation of the intestinal lining as seen in genetically predisposed patients (those with the DQ2/8 molecules) [15].
Sensitivity to gluten is not restricted to humans; varying forms of gluten-dependent enteropathies have been reported in rhesus macaques [16], dogs [17], horses [18], and mice [19]. Although, gluten sensitivity has not been reported in monogastric pigs and poultry, several studies have evaluated its effect as an ingredient in feed. Most of these studies have focused on the effect of wheat gluten on performance [20–22], involvement in disease [23] and gut health [24–26]. To the best of our knowledge, no study has been done on the impact of gluten on the physiology of the chicken intestinal tract.
RNA-Sequencing is a sequencing methodology which uses next-generation sequencing to reveal the presence and quantity of mRNA in a biological sample, analyzing the continuously changing cellular transcriptome [27]. This method can detect transcripts from any organisms with previously undetermined genomic sequences and allows for accurate quantification of RNA. So, this study therefore aimed to study the impact of a high gluten diet on intestinal morphology of starter and grower broilers as well as the host response to such a diet through transcriptomic techniques (RNA-seq). We hypothesized that a gluten-rich diet affected intestinal development differently between young (1-week-old) and older (4-week-old) chicks.
MATERIALS AND METHODS
This study was approved by the Animal Ethics Committee of Jeonbuk National University, Korea (CBNU-2017-00101).
A total of 120 one-day-old broilers (Ross Strain) were used in this study (Dongwoo Hatchery, Iksan, Korea) to employ a two-level general factorial design to investigate the gluten intervention by bird age. As described in the Supplementary Fig. S1, birds were divided into 4 groups by different gluten feeding age (1 week or 4 weeks) and gluten inclusion level (0% or 25%). The broilers were allowed ad libitum access to water and feed under light on all days. Starter and grower broiler feeds were purchased from TS Corporation (Seoul, Korea) which compositions were reported in our previous study [28], and pure wheat gluten was obtained from Singsong Food (Nonsan, Korea). During gluten feeding challenges, the control group was allowed access to a 100% broiler feed (this was considered the wheat gluten-free diet) while the gluten-fed group was allowed a feed containing 25% gluten (i.e., 75% broiler feed + 25% gluten supplement). At the end of each gluten challenge, 6 birds (3 controls and 3 gluten-fed) were randomly selected and euthanized by cervical decapitation and duodenum samples collected, respectively. A total of 12 samples for histological examination were fixed with 10% neutral buffered formalin (NBF) while samples for RNA sequencing were immediately frozen in liquid nitrogen and stored at −70°C until analysis. Birds and feed were weighed to calculate feed intake and body weight during gluten feeding challenges to compare growth performance among groups.
Fixed duodenum samples from the experimental chickens were sectioned and embedded into paraffin blocks. The paraffin-embedded tissue blocks were sectioned at 5 μM, de-paraffinized in xylene and hydrated in a descending series of ethanol and then stained with hematoxylin and eosin (HE) for histological examination. Stained sections were dehydrated in an ascending series of ethanol, cleared in xylene and mounted on slide. Digital images were acquired using a Leica DM2500 microscope (Leica Microsystems, Wetzlar, Germany) at fixed 100× magnification.
RNAs were isolated from duodenum mucosal samples from 12 chickens using TRIzol reagent (Invitrogen, Waltham, MA, USA) for constructing RNA-seq libraries using TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s guidelines. Library quality (size distribution and concentration) was checked using Agilent Technologies 2100 Bioanalyzer. In order to check library quality, we loaded 1µL of sample on DNA 1000 Bioanalyzer LabChip and selected 150–200 bp size. Libraries with different indexes were pooled and sequenced in one lane using Illumina NextSeq 500 high-throughput sequencing instrument with 75 paired-end (PE) reads. An average of 17.01 million sequencing reads per sample were generated. Sequencing data were deposited at Sequence Read Archive (SRA) with project number PRJNA488337.
After obtaining RNA-sequencing data, raw reads were preprocessed for quality control using Trimmomatic-0.36 (trimming read and LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36). Subsequently, we obtained chicken reference genome (Gallus_gallus-5.0) from Ensemble database. We performed the indexing process based on the chicken reference genome. Reads were aligned with this chicken genome using HISAT2 (version 2.1.0) [29], a fast and sensitive alignment tool for mapping next-generation sequencing reads. The aligned sequence reads and mapping rate of small intestine samples from all twelve chickens are presented in Supplementary Table S1. These aligned chicken sequence reads were then used for downstream analyses of host differential expression.
After we obtained chicken genome GTF file (Gallus_gallus-5.0.9) from the Ensemble database, we counted the number of reads for each gene using FEATURECOUNT [30] (version 1.5.3) within the subread aligner packages [31]. Based on the assumption that the number of RNA-Seq reads produced by a transcript is directly proportional to its abundance, we measured gene expression level based on read count.
In this study, we used DESeq2 (version 1.18.1) [32], a package in R [33] to identify differentially expressed genes (DEGs) between gluten-fed and non-gluten-fed broilers at 1 week and 4 weeks of age. To enhance the statistical power for identifying DEGs, we removed genes with weak expression levels using HTSFilter package [34]. First, DESeq2 used empirical Bayes shrinkage method to estimate dispersions and fold changes by modeling read counts following a Negative Binominal distribution. Then Wald test p-value was inferred to evaluate the statistical significance. The estimated p-values were adjusted using the False Discovery Rate (FDR) method to control for multiple hypothesis testing as is typical in RNA-Seq datasets. Finally, DEGs were declared at a significant level of |log2 (fold change) | > 2 and FDR < 0.05. Chicken Ensemble gene IDs were converted into official gene symbols by cross matching to chicken and human Ensemble gene IDs. And then, we used several plotting methods (density plot, scatter plot and principal component analysis [PCA] plot) to identify the quality of RNA-seq as well as relationships between non-gluten and gluten-fed chickens using DESeq2 [32], ggplot2 [35] and affy [36] R packages. We also performed a heatmap clustering analysis was performed on the DEGs using ggplot2 [35] in order to further identify expression patterns among the groups. The official gene symbols of chicken genes were then used for functional clustering and enrichment analyses using WebGestalt [37]. Representative functional groups in the compared treatments (gluten and non-gluten fed chicken) at 1 week of age were investigated using gene ontology (biological process, cellular component, molecular function) information and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. To identify enriched pathways, functionally clustered genes were clustered using overrepresentation enrichment analysis (ORA) method and KEGG pathway database.
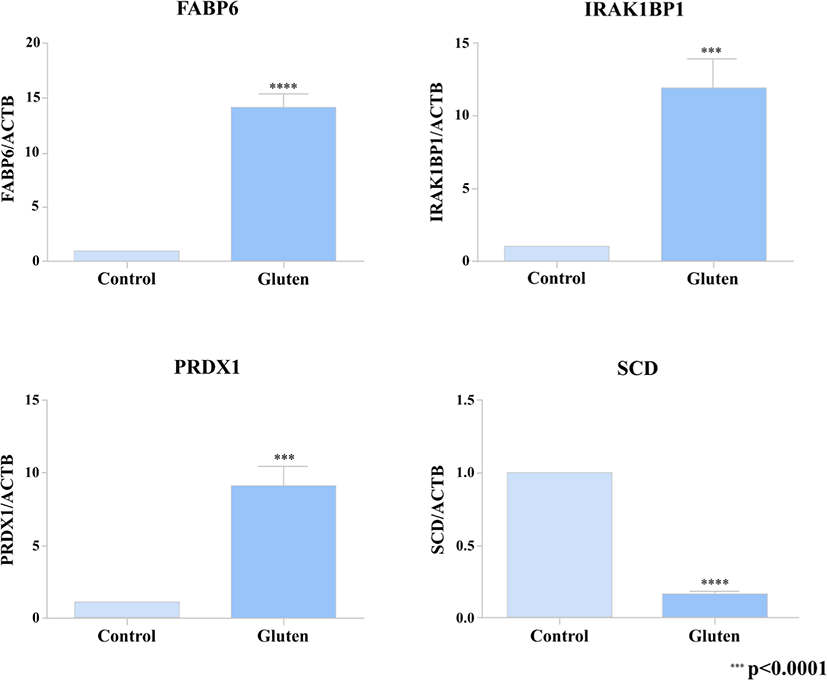
In order to verify the DEG’s from our transcriptome analysis (RNA-seq data), we used a quantitative real-time PCR to detect the expression pattern of 4 of the DEG’s between the gluten-fed and control groups at 1 week of age. Two of the selected genes (FABP6, SCD) were associated with a KEGG pathway whose differential expression showed statistical significance and the other 2 DEG’s (PRDX1 and IRAK1BP1) were randomly selected. For the qRT-PCR, we used samples from 6 one-week old broilers (three individuals from the control group and three individuals from the gluten-fed group). The primers used are presented in Supplementary Table S2. To synthesize cDNA, ReverTra Ace (Toyobo, Osaka, Japan) was used according to manufacturer’s instructions. Real-time qPCR was performed using iTaq Universal SYBR Green Supermix (Bio Rad, Hercules, CA, USA) according to the manufacturer’s instructions. PCR reaction consisted of the following: 1 μL of diluted cDNA, 5 μL of iTaq Universal SYBR Green Supermix, and 1 μL each of 5 pmol/μL diluted forward and reverse primers. C1000 Thermal Cycler (Bio Rad) was used to measure the expression level of target gene. Conditions used for real-time qPCR were as follows: incubation at 95°C for 5 minutes followed by 40 cycles of denaturation at 95°C for 10 seconds and annealing at 64°C for 30 seconds. All measurements were performed three times for each sample. Relative gene expression was calculated using the 2–ΔΔCt method [38]. Relative expression of the target gene was normalized to that of β-actin (ACTB).
Results are shown as mean with standard error bar. All data were analyzed by a T-test procedure using the R statistical program [33]. Statistical significances were considered at p < 0.05, p < 0.01 and p < 0.0001.
RESULTS
We performed a gluten feeding experiment (gluten: 0% or 25%) on chicken at 1 week old and 4 weeks of age. Chicken fed a non-gluten diet showed higher feed intake and body weight gain during gluten feeding challenge both at 1 week and 4 weeks of age (p < 0.05) (Supplementary Tables S3 and S4).
Quality filtration of the RNA-seq data using the Trimmomatic tool left 95.35% good quality RNA-seq reads from the samples to be used for downstream analysis. Average numbers of sequence reads were 16.1, 17.4, 16.8, and 17.7 million for the non-gluten-fed 1-week-old group, 25% gluten-fed 1-week-old group, non-gluten-fed 4-week-old group, and 25% gluten-fed 4-week-old groups respectively. The rates of alignment for reads from these 4 groups mostly exceeded 94%. Reads were successfully mapped to the chicken reference genome (Gallus_gallus-5.0) using Hisat2 [29]. The sequence read order, sequence yield, total number of reads and mapping rates for each sample are shown in Supplementary Table S1. Further evaluation and comparison of the RNA-seq data from the small intestine tissue samples was achieved through visualizations such as the density plot, correlation plot, and PCA plot (presented in the Supplementary Figs. S2 and S3).
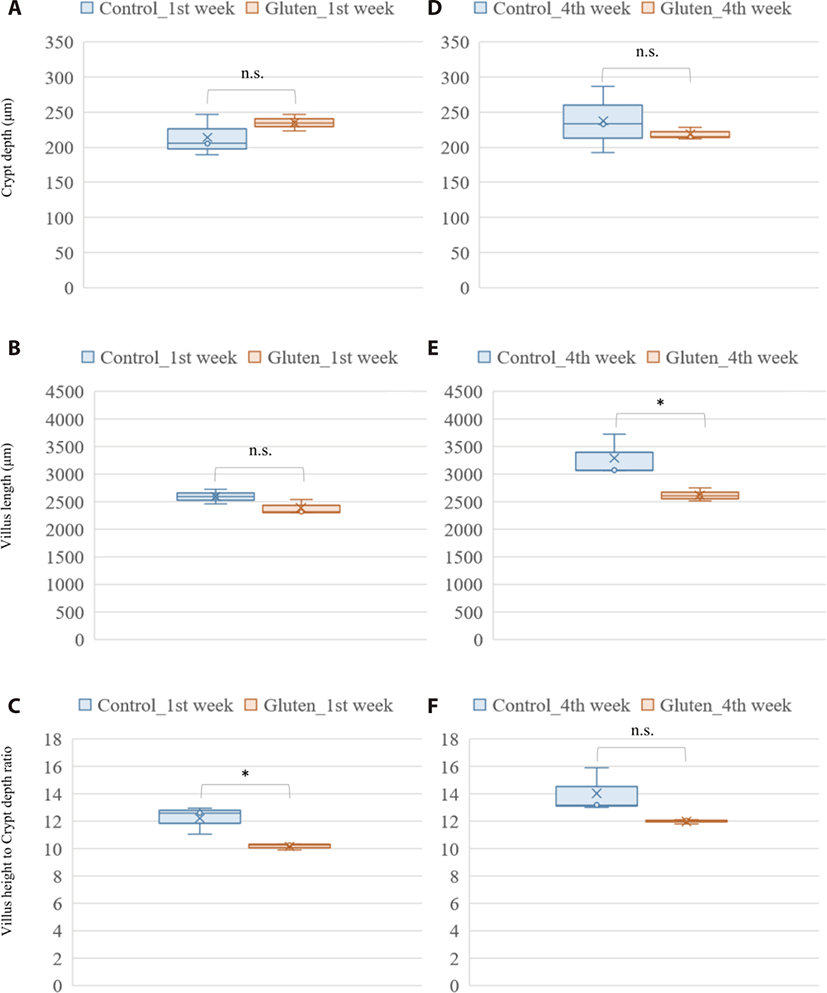
Photomicrograph analysis of the layers of duodenal mucosa revealed the effect of a high gluten diet on intestinal morphology in broilers (Fig. 1). In the crypt depth, gluten feeding showed no significant difference in both ages. Gluten-feed broilers had a lower villus height in the 4-week-old of age (p-value = 0.044). Collectively, the villus height to crypt depth ratio was lower in the 1-week-old, gluten fed broilers (p-value = 0.027).
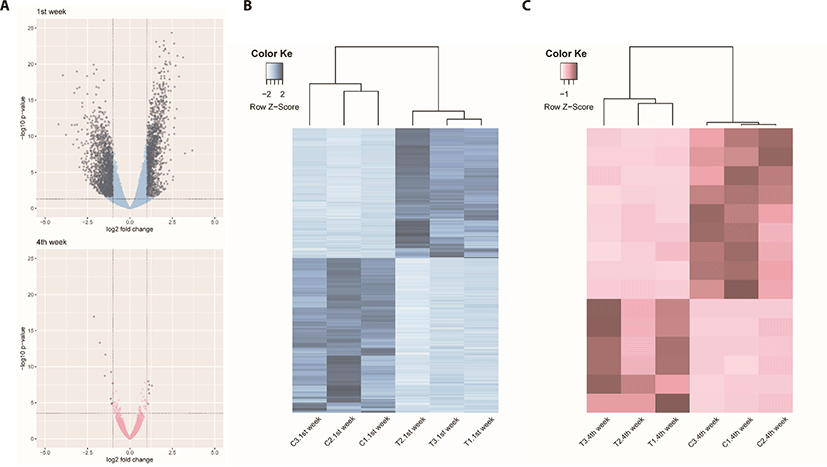
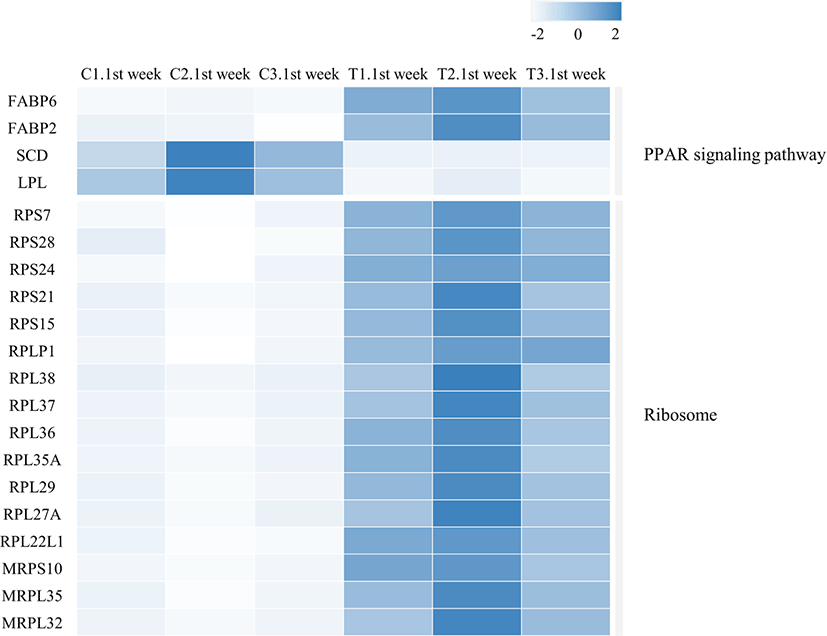
We identified 3,041 DEGs at 1 week and 15 DEGs at 4th week by the criterion (|log2FC| > = 1). However, we identified 289 DEGs at 1 week, but only one DEG week 4 by the criterion (|log2FC| > = 2). The number of DEGs between the gluten-fed and control groups of 1-week-old broilers was 289 (91 upregulated, 198 downregulated) (|log2FC| > = 2) (Supplementary Table S5). At 4 weeks of age, the number of DEGs in the gluten-fed group relative to the controls was one (downregulated) and this gene was not shown in DEGs of 1 week old group. Expression levels of significant DEGs in 1-week-old and 4-week-old chicken groups are shown in Fig. 2. Of the 289 DEGs found between the gluten-fed and control groups of 1-week-old chickens, FABP6 gene had the highest base mean read count and the most significant (Supplementary Table S5). This gene encodes one of the small ubiquitous cytoplasmic proteins, FABP6, that can bind hydrophobic ligands including long-chain fatty acids. This gene is involved in the uptake, transport and metabolism of fatty acids as well as in the peroxisome proliferator-activated receptor (PPAR) signaling pathway.
We identified the top 10 biological pathways related to DEGs between the two groups (25% gluten-fed and control groups) at 1 week of age based on the KEGG pathway analysis. Among the enriched pathways, Ribosome, Oxidative phosphorylation, PPAR signaling pathways showed statistically significant enrichment among the treatment group (Table 1). In gene ontology analysis, we used three functional databases; biological process, cellular component and molecular function. We obtained 10 biological processes, 10 cellular component terms and 11 molecular functions among the gene ontologies of the DEGs in gluten fed and control chicks at 1 week of age (Table 2). In biological process gene ontology analysis, identified DEGs were most relevant to substrate adhesion-dependent cell spreading and ribosome assembly. Additionally, 9, 7, 7 were strongly associated with cellular component morphogenesis, translation, and peptide biosynthetic process, respectively. In cellular component gene ontology analysis, 7 genes were commonly relevant to cytosolic ribosome, ribosomal subunit, ribosome, and cytosolic part. These 7 genes plus another gene, MDN1 were associated with intracellular ribonucleoprotein complex and ribonucleoprotein complex. In molecular function part of DEGs gene ontology analysis, structural constituent of ribosome and structural molecule activity were the most significant and metal ion binding was relevant to numerous DEGs (15 genes).
Also, we selected 2 DEGs (FABP6, SCD) associated with results of KEGG pathway analysis and randomly extracted 2 DEGs (PRDX1 and IRAK1BP1) to compare qRT-PCR validation with RNA-seq results. A high correlation was found between results from RNA-seq data and the levels of expression detected using the PCR (Fig. 3).
Unlike comparison at 1 week age, we adopted the criterion (|log2FC| > = 1) to compare the DEGs between two growth stages by gluten feeding in 4-week-age chicken data analysis. The number of DEGs between the gluten-fed and control groups of 4-week-old broilers was 15 (6 upregulated, 9 downregulated) (Supplementary Table S6). We identified 10 gene functions out of 15 DEGs in 4-week age chickens compararison (between control and gluten-fed)
DISCUSSION
In this study the effect of a high gluten diet on small intestines of broiler chicks was investigated. We evaluated the response of chicken duodenum both histologically and in gene expression using transcriptomic techniques. We hypothesized that feeding chicken a high gluten diet would result in damage to the physiology and immune function of the small intestines with consequent retardation in intestinal development. This is the first report that we know of describing the gene expression pattern associated with high gluten diet in broilers. We identified a total of 289 DEGs between broiler chicks fed a high gluten diet (25% gluten) and those fed a control diet (0% gluten) at 1 week of age and only one DEG when comparing the groups at 4 weeks of age. In addition, PCA plots of the read count data from the host gene expression, clearly distinguished between high-gluten-fed and control groups of at 1 week of age. However, the PCA plot did not distinguish between the high-gluten-fed and control groups at 4 weeks of age (Supplementary Fig. S3). This was surprising because our results had revealed damage to the duodenal mucosa in villus height as well as a reduction in feed intake and weight gain among the gluten-fed groups at all ages within the study. We speculate that damage may not be significant to detect the DEGs in aspect of the collective indicator of villus height to crypt depth ratio between control and gluten-fed broilers at 4 weeks of age.
The effect of a high-gluten containing diet on growth performance of the broilers in our study was consistent with findings from previous studies on not only diets rich in wheat gluten [20] but also diets rich in corn gluten [39]. In the current study, we found a significant decrease in feed intake and average daily gain among broilers fed a diet with high inclusion levels of wheat gluten. In their study, Afshar and Moslehi [20] argued that consumption of feed with high proportions of gluten raised the viscosity of intestinal content due to the water absorbing property of gluten. And that it was this high viscosity that led to a reduction in feed intake with a subsequent impact on weight gain [20]. Also, previous study about clinical improvement in response to a gluten-free diet in human celiac disease patient performed intestinal biopsy to examine characteristic histopathologic changes of the duodenum. Celiac disease is a serious autoimmune disease that occurs in genetically predisposed people where the ingestion of gluten leads to damage in the small intestine. This previous study could find abnormal endoscopic and histopathologic appearances in the majority of patients with celiac disease (pubmed.ncbi.nlm.nih.gov/12556782/). Histologically, the gluten rich diet induced a significant reduction in villus height within the duodenum of chicken in the treatment group at 4 weeks of age in this study. This was in agreement with previous studies that found wheat gluten induced reduction in villus height [28,40]. Further analysis of the photomicrograph also revealed a reduction in the ratio of villus height to crypt depth among the gluten-fed broilers, especially at 1 week of age. A reduced villus height to crypt depth ratio is associated with reduction in absorptive potential of the small intestine [41,42]. In regard to these gluten-induced effect on villi architecture, the observed reduction in body weight gain among the gluten-fed broilers relative to the controls is not a surprise (Supplementary Tables S3 and S4).
There is a possibility that the revealed changes in gene expression at 1 week were involved in the manifestation of histological changes within the duodenal mucosa and subsequent drop in feed intake and body weight gain of the gluten fed birds. We therefore examined the DEGs by performing KEGG pathway and gene ontology analyses in order to get insights into their role in the observed effects of a high gluten diet. A notable finding from this study was, that the expression of genes involved in the ribosome pathway were significantly up regulated in the duodenal mucosa of chicken fed a high gluten diet. Sixteen (16) DEGs were associated with ribosome protein biogenesis and ribosome assembly. Thirteen of these DEGs, (RP S7, RPS28, RPS24, RPS21, RPS15, RPLP1, RPL38, RPL37, RPL36, RPL35A, RPL29, RPL27A and RPL22L1) coded for cytosol ribosomal protein, and three genes (MRPS10, MRPL35 and MRPL32) coded for mitochondrial ribosomal protein. These proteins play an important part in the assembly of the ribosomal structure and enabling the translational role of the ribosome. However, the up regulation of these genes would lead to the increased expression of 16 out of approximately 79 ribosomal protein in chicken cells [43]. It is known that a dysregulation of ribosomal proteins is a feature of ribosomal stress [43,44]. Among the triggers of ribosomal stress are; physical, chemical and irradiation agents that perturb ribosomal protein biogenesis as well as lack of nutrients [43]. The elastic gluten proteins have been shown to increase digesta viscosity within the chicken gut which in turn affects gut movement, and nutrient absorption [45]. It is therefore plausible that these physical characteristics of gluten in the gut might trigger a ribosomal stress. The observed effect on the villi-crypt arrangement in this study could also account for an interruption of nutrient absorption. An unbalanced increment in the ribosomal proteins induces their regulation through the activation of p53 in the MDM2/MDMX-p53 cascade [46] which in turn results in arrest of the cell cycle and consequently, cell proliferation. Indeed, some of the upregulated ribosomal proteins; RPL37, RPS7 and RPS15 are known to bind to MDM2/X thus preventing its degradation of p53 [47,48]. We speculate that it is this ribosomal protein induced accumulation of p53 that causes the retardation of villi development observed histologically among the gluten fed birds.
Another significantly upregulated pathway was the PPAR signaling path. This pathway includes a collection of receptor proteins found in the nucleus which, as transcription factors, regulate expression of certain genes. Their functioning is critical to cellular differentiation, development, and lipid metabolism [49]. Four genes within this pathway were differentially expressed; FABP2, FABP6, LPL and SCD (Table 1 and Fig. 4). Of these, FABP2 and FABP6 were upregulated while LPL and SCD genes were downregulated in the duodenal samples from high-gluten-fed broilers. The FABP6 gene codes for the Fatty acid Binding Protein 6 (also known as the ileal FABP/ Il-FABP). The protein plays a role in the uptake and transport of bile salts into the enterocytes [50]. Therefore, the upregulation of this gene would result in the increased production of the FABP6 protein leading to increased uptake of bile salts into the enterocytes. Accumulation of bile salts into the enterocytes has been shown to induce apoptosis and/ arrest of cell growth [51]. It is therefore plausible that upregulation of this gene caused the observed retardation of villi development in the duodenum of chicken fed a high gluten diet. FABP2 codes for the intestinal FABP protein (also known as FABP2/ I-FABP). This binding protein is involved in the uptake and transport of the fatty acids [52]. The FABP2 protein carries the fatty acid towards the triacylglycerol pathway leading to accumulation of triacylglycerols and cholesteryl [53] which are then secreted into circulation via the intestinal lymphatic system. The SCD gene codes for Stearoyl-coenzyme A desaturase (SCD1) which mediates the synthesis of mono-unsaturated fatty acids (MUFAs). One of these MUFAs, Oleic acid, is a building block for the complex lipids that support cell proliferation [54]. In addition, oleic acid is an endogenous ligand of the PPAR-γ which is an important transcription factor in anti-inflammatory signaling [55]. Therefore, the down regulation of the SCD gene by the high gluten diet would lead to the suppression of the anti-inflammatory activity of PPAR-γ. Indeed, it has been reported that SCD ablation in mice promotes inflammation [56]. This is also in agreement with findings from our previous study where we reported a chronic inflammation in the duodenal mucosa of 25% and 50% gluten fed broilers [28]. In the study, the histological signature of chronic inflammation was supported by an upregulation of inflammatory mediators (tumor necrosis factor [TNF]-α, interleukin [IL]-12, IL-8 and IL-6) [28]. This implies that a gluten-induced downregulation of SCD in the current study resulted in a shift of balance in the modulation of inflammation with a resultant enabling of a chronic inflammatory process in the chicken duodenum. In addition, SCD promotes cholesterol and phospholipid production and consequently, promotes membrane lipid production [57]. Downregulation of this gene would therefore reduce membrane formation and eventually suppress cell proliferation. The LPL gene codes for the lipoprotein lipase (LPL) enzyme. The enzyme is produced in several tissues including the duodenum but has its physiological role in the luminal surface of endothelial cells of capillaries. LPL catalyzes the hydrolysis of plasma triacyl glycerides TAG’s into free fatty acids and monoacylglycerols for tissue utilization (reviewed by Alpers et al. [58]). Therefore, a downregulation of this gene in the duodenum, would theoretically results in reduction of LPL activity in the duodenum and as a result a reduced uptake of fatty acids from plasma into the duodenal tissues. However, LPL is produced in multiple sites and its regulation can be tissue specific (reviewed by Braun and Severson [59]). This therefore may not mean a systemic reduction in plasma triglyceride rich lipoproteins or a consequential systemic increase in fat storage/deposition in the broilers. In contrast, the LPL gene is upregulated in gluten-induced enteropathy among celiac patients [60] but was downregulated in our gluten-fed broilers. Further, the PPAR signaling pathway is downregulated in human celiac patients [61,62] and a gluten-free diet alleviates signs, at least in part, by upregulation of the pathway [62]. Considering our findings in the PPAR signaling pathway modulation, it could be inferred that a high gluten diet promoted the transport of triacylglycerols (storage form of lipids) across the enterocytes while lowering fatty acid uptake from circulation and therefore utilization in the duodenal tissues. This would further explain the retarded development of duodenal villi observed in the broilers fed a high gluten diet in our study since fatty acids are crucial to cell proliferation as structural and functional components.
The third pathway that was significantly differentially expressed was the oxidative phosphorylation pathway. Seven genes in this pathway (NDUFB1, NDUFB7, NDUFV3, UQCR10, COX17, ATP5J and ATP5H) were upregulated in the duodenum of broilers fed a high gluten diet. These genes code for subunits of the complexes I, III, IV and V of the mitochondrial electron transport chain (ETC). Three of these genes, NDUFB1, NDUFB7 and NDUFV3, code for accessory subunits of the mitochondrial complex I (NADH: ubiquinone oxidoreductase). Very little is currently known about the particular roles played by these accessory subunits. However, the NDUFB1 subunit is one of the single transmembrane domains (STMD) thought to stabilize and therefore promote the assembly of the transmembrane helices of the hydrophobic core subunits of complex I within the mitochondrial membrane [63]. The NDUFB7 subunit is critical to the proton pumping function of the mitochondrial complex I. Without it, the entire distal proton pumping module (PD module) is disabled and about half of the proton pumping function of complex I is lost [64]. Both NDUFB1 and NDUFB7 are components of the PD module of complex I. Enhanced proton pumping across the inner mitochondrial membrane leads to generation of a proton motive force which has been implicated in the increased production of reactive oxygen species by complex I [65]. On the other hand, the NDUFV3 subunit is a structural component of the electron input module (N module). UQCR10 codes for a subunit of complex III (Coenzyme Q-cytochrome c oxidoreductase) that plays a role in the assembly of complex III of the ETC [66] and therefore contributes to the reduction of cytochrome c. COX17, codes for the Cytochrome c oxidase copper chaperone protein which is a subunit of complex IV (Cytochrome c oxidase). This protein plays a vital role in the assembly of as well as recruitment and presentation of copper to the mitochondrial cytochrome c oxidase [67]. Two of the upregulated genes (ATP5J and ATP5H) in this pathway code for subunits of complex V (F0F1 ATP synthase) of the ETC. ATP5J codes for the ATP synthase, H+ transporting mitochondrial F0 complex subunit F6 that is responsible for connecting the lipid-soluble inner membrane-bound F0 region to the water soluble, matrix-exposed F1 region of the complex[68]. ATP5H codes for the ATP synthase, H+ transporting mitochondrial F0 complex subunit D whose putative role is in the assembly of ATP synthase [69]. Overall, increased oxidative phosphorylation is a feature of non-proliferating or quiescent cells, given that cell proliferation is skewed more towards aerobic glycolysis than oxidative phosphorylation (as reviewed by Vander Heiden et al. [70]) However, it remains unclear whether these subunits work in-sync with the other subunits of the mitochondrial complexes they belong to or whether they could function independently within the cytoplasm and/or mitochondrial matrix. And in the latter case, which of their roles would be influenced greatest by their dysregulation. In 15 DEGs from 4-week age chickens compararison (between control and gluten-fed), we could find gene function of 10 genes (Supplementary Table S6). Among them, we confirmed two down-regulated genes (MGAM, MLXIPL) related to carbohydrate metabolism and one up-regulated gene (PDK4) in lipolysis due to an increase in gluten intake. In addition, we were able to confirm the decrease in the expression of one immune protein gene (DMB1) and observed two genes (DFNA5, IFI6) related to apoptosis in two small intestine tissues. Also, we confirmed the increase in the expression of one intestinal inflammation gene (SAMD9L), and interestingly, observed that the CDCP2 motif-related gene (CDCP2; CUB domain containing protein 2) showed a difference in expression. The CUB3 domain is a widely occurring structural motif, and this protein is involved in many biological functions (including tissue repair, cell signaling). The decreased gene expression of CDCP2 motif proteins in the gluten-fed group suggested that gluten could induce widespread biological function decline.
Additionally, we thought that transcriptomic comparisons and following DEG analyses between 1 week and 4 weeks per normal (1 week vs 4 weeks in normal) and treatment (1 week vs 4 weeks in treat), respectively can identify transcriptomic changes between ages in two different aspects. We could identify 354 DEGs between 1 week and 4 weeks in control and 3,591 DEGs between 1 week and 4 weeks in gluten treatment (Supplementary Figs. S4–S7). Based on this result, we found that gluten supplementation induces many trancriptional changes.
In conclusion, this study presents the transcriptomic profile of the duodenum in broilers fed a high gluten containing diet. Our findings suggest that a high gluten diet induces an upregulation of genes involved in ribosome biogenesis, PPAR signaling and oxidative phosphorylation in 1-week-old broilers. Deductions from this pattern indicate that high gluten in the diet causes chronic inflammation and transcriptional inhibition of cell proliferation within the duodenal mucosa. However, the gluten-induced transcriptional changes were more pronounced early in the life of the broilers (1 week of age) but not at a later age (4 weeks of age). It is unclear why we were unable to detect significant transcriptional changes at 4 weeks of age despite the histological change of the villus height observed. However, it is plausible that a reduced transcriptional response might be, in part, a coping mechanism that boosts the birds’ resilience towards the nutritional stress due to a high gluten diet [71]. This study was limited by a small RNA-seq sample size [72] and the inherent difficulty of interpreting transcriptome analysis involving multiple cell type (in this case, the various cell types within the duodenum). However, in the right context, our findings show that our approach could simultaneously characterize the diversity of the differential expression of several genes from the host in response to gluten-feed. It has a direct and broad applicability in nutrigenomic and veterinary studies. Despite the insights from this study, many questions remain. More needs to be understansd about the roles played by these DEGs so that more reliable/conclusive deductions can be made about such observed patterns in transcriptional responses.
