INTRODUCTION
The global demand for functional animal feed additives is increasing rapidly, driven by the need to enhance animal health and promote agricultural sustainability [1]. High-calorie grain-based diets, commonly used to improve feed efficiency in livestock, have been associated with issues such as immune disorders and Escherichia coli O157 infections in cattle [2]. Furthermore, the rising costs of grain imports, which constitute a significant portion of feed expenses worldwide, underscore the urgent need for alternative solutions [3,4]. Consequently, developing feed additives that can reduce grain dependency while enhancing nutritional value and feed efficiency, particularly in fiber-based diets, has emerged as a pressing priority [5].
Xylo-oligosaccharides (XOS) are well-established functional oligosaccharides that act as prebiotics, promoting the growth of beneficial gut bacteria such as Bifidobacterium and Lactobacillus, thereby improving gut health, enhancing immune function, and increasing digestive efficiency in animals [6]. XOS can be derived from xylan, a dietary fiber, through the action of endo-xylanase [7]. Similarly, yeast protein, rich in vitamin B and high-quality protein, serves as a valuable nutritional source, contributing to disease prevention, metabolic balance, and improved feed efficiency [8]. The inclusion of XOS and yeast protein in feed additives not only enhances their nutritional value and feed efficiency but also provides a sustainable and cost-effective alternative to expensive antibiotics and protein supplements.
Corn cobs are a major by-product of corn production, with millions of tons discarded annually [9]. They consist of 30%–40% hemicellulose, 35%–45% cellulose, and 5%–20% lignin, with a notably high content of xylan-based hemicellulose (35%–40%). This composition makes corn cobs a promising lignocellulosic biomass resource for producing high-value products such as XOS [10]. Globally, there is a growing interest in utilizing agricultural residues as renewable resources for food and biofuels. Corn cobs are relatively inexpensive and widely available, positioning them as an ideal raw material for developing sustainable feed additives [6]. Furthermore, while corn cobs contain various nutrients, such as beta-sitosterol and minerals, their digestibility is low when fed directly to animals. As a result, microbial fermentation of corn cobs to produce feed presents an effective alternative [11].
Although advancements have been made in the production of XOS and yeast protein from lignocellulosic biomass, several challenges remain. Conventional methods often rely on extensive chemical treatments or multi-step enzymatic processes, which are complex, costly, and associated with significant environmental impacts [12]. Some studies have successfully produced XOS and yeast protein using reed sawdust as a substrate; however, these processes remain complex, as XOS and yeast protein are produced separately [13]. This underscores the need for simplified and environmentally sustainable approaches to produce XOS and yeast protein using agricultural by-products simultaneously.
This study aimed to develop an integrated and environmentally sustainable process for producing a functional feed additive enriched with XOS and yeast protein from corn cobs. To achieve efficient XOS production, a recombinant Saccharomyces boulardii strain expressing the endo-xylanase gene on its cell surface was employed. A simultaneous saccharification and fermentation (SSF) process was conducted using pretreated corn cobs under various conditions, combining the recombinant strain with commercial cellulase. This process enabled the concurrent production of XOS and yeast protein by efficiently utilizing the xylan and cellulose present in corn cobs. To the best of our knowledge, this study represents the first successful attempt to simultaneously produce XOS and yeast protein using a recombinant S. boulardii strain engineered to express endo-xylanase on its cell surface. This innovative approach highlights the potential for developing cost-effective and environmentally sustainable functional feed additives by repurposing agricultural residues. Furthermore, this strategy aims to enhance the nutritional value and digestibility of fiber-based diets, thereby providing a robust foundation for the development of sustainable feed solutions that reduce dependence on grain-based feed.
MATERIALS AND METHODS
Corn cobs were obtained from Tojongherb and used as the substrate for an integrated process to produce XOS and yeast protein. A commercial cellulase (Cellic® Ctec3) was purchased from Novozymes and used to hydrolyze the residual cellulose in the pretreated corn cob into glucose, which served as a nutrient source for yeast growth. Glucose and xylose (X1) were procured from Sigma-Aldrich. Beechwood xylan, xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5) and xylohexaose (X6) were obtained from Megazyme.
The recombinant yeast strains and plasmids used in this study are detailed in Table 1. Yeast cells were pre-cultivated in yeast extract and peptone (YP) medium (10 g/L yeast extract and 20 g/L peptone) supplemented with 20 g/L dextrose (YPD). The pre-cultivation was performed at 30°C and 250 rpm for 24 h, after which the cells were used in subsequent experiments. E. coli DH5α (New England Biolabs) was used for amplifying gRNA plasmids. E. coli cells were cultivated in luria-bertani (LB) medium (5 g/L yeast extract, 10 g/L tryptone, 10 g/L NaCl) supplemented with 100 μg/mL ampicillin (LBA). Cultivation was conducted at 37°C and 250 rpm for 18 h.
| Strains | Description1) | References |
|---|---|---|
| Saccharomyces boulardii | ||
| SB | Wild-type; Saccharomyces boulardii (ATCC MYA-796) | [33] |
| X-Aga2 | SB int#1::PCCW12-MFα1-TsaGH11-AGA2- TCYC1 | This study |
| X-Cwp1 | SB int#1::PCCW12-MFα1- TsaGH11-CWP1- TCYC1 | This study |
| X-Cwp2 | SB int#1::PCCW12-MFα1- TsaGH11-CWP2- TCYC1 | This study |
| X-Sed1 | SB int#1::PCCW12-MFα1- TsaGH11-SED1- TCYC1 | This study |
| X-Pir1 | SB int#1::PCCW12-MFα1- TsaGH11-PIR1- TCYC1 | This study |
| X-Tir1 | SB int#1::PCCW12-MFα1- TsaGH11-TIR1- TCYC1 | This study |
| Plasmids | ||
| pRS41N-Cas9 | pRS41N plasmid containing a natNT marker and Cas9 | [46] |
| pRS42H-cg#1 | pRS42H plasmid containing a hph marker and gRNA for the cg#1 site | [47] |
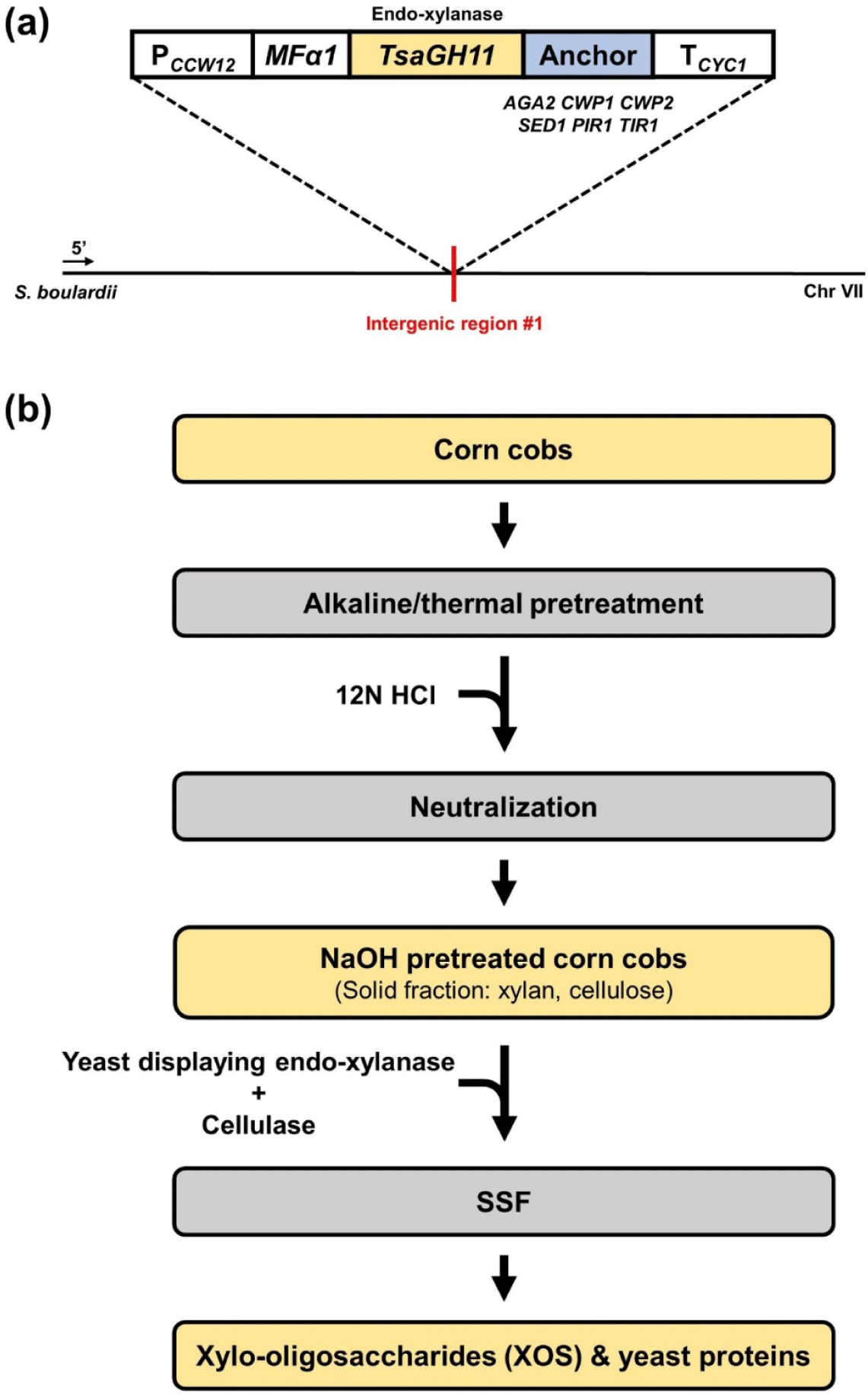
The yeast transformation process followed a previous study [14,15], with the following modifications applied in this study (Table 2). To construct expression cassettes for endo-xylanase, the TsaGH11 gene (UniProt: I3VTR8) from Thermoanaerobacterium saccharolyticum was codon-optimized for E. coli [16]. The endo-xylanase gene, fused with six different anchor protein genes derived from S. boulardii, was integrated into the cg#1 sequence of the S. boulardiiPCCW12-MFα1-cg#1-TCYC1 strain, which had been previously constructed [15], using the pRS42H-cg#1 gRNA plasmid. Finally, six strains expressing endo-xylanase with different anchor proteins, X-Aga2, X-Cwp1, X-Cwp2, X-Sed1, X-Pir1, and X-Tir1, were constructed (Fig. 1A).
To compare the enzymatic activity of endo-xylanase anchored on the cell surface, the experiment was conducted under the following conditions, using the methodology described by Kim et al. [16] with modifications: Cells were cultivated in YP medium supplemented with 20 g/L glucose (YPD) until reaching an exponential-phase cell density of 0.5 g DCW/L. The cells were then harvested by centrifugation at 3,500×g for 5 min and washed twice with sterile distilled water. The experiment was conducted with a cell concentration of 20 g DCW/L in a reaction mixture containing 0.5% (w/v) beechwood xylan as the substrate and 50 mM sodium acetate buffer (pH 5.0) in a total reaction volume of 1,000 µL. The reaction was carried out at 30°C and 130 rpm for 30 min. To terminate the reaction, the cells were centrifuged at 20,000×g for 1 min, and a 40 μL aliquot was taken. The enzyme reaction was halted by adding 80 μL of 3,5-dinitrosalicylic acid (DNS) reagent, followed by heating at 95°C for 5 min to allow color development. After centrifugation at 20,000×g for 10 min at 25°C, 100 μL of the supernatant containing the released sugars was collected, and absorbance was measured at 540 nm using a SpectraMax Pro iD3 microplate reader (Molecular Devices). All experiments were performed in biological triplicates.
Fermentation was performed in a 100-mL flask containing 20 mL of YP medium supplemented with 20 g/L glucose and 20 g/L xylan solution (YPDX) at 30°C and 130 rpm. The initial optical density (OD) was adjusted to 1. All experiments were performed in biological triplicates.
Corn cobs were ground into particles smaller than 1.00 mm using a mechanical grinder. For pretreatment, 5 g of the ground powder was mixed with 45 mL of one of the following solutions: distilled water (DW), 1% (w/v) sulfuric acid (H₂SO₄), 2% (w/v) sodium hydroxide (NaOH), 7% (w/v) NaOH, or 12% (w/v) NaOH solution, maintaining a solid loading of 10%. The mixtures were subjected to thermal treatment at 121°C for 20 min. Following heat treatment, the pH was adjusted to 6.5 by adding 12 N hydrochloric acid (HCl) to neutralize the samples.
The neutralized samples were centrifuged at 3,500×g for 20 min to recover the solid fractions, and the supernatant was discarded. The recovered solid fractions were subjected to lyophilization and used in subsequent fermentation experiments (Fig. 1B).
The pretreated corn cobs were subjected to SSF using the previously selected X-Tir1 strain and a commercial cellulase. Fermentation was initiated by adding the X-Tir1 strain and Cellic® CTec3 cellulase (3% v/v; mL-Cellic® CTec3/g-biomass) to a YP medium (15 g/L yeast extract, 30 g/L peptone) containing the pretreated corn cobs at a 10% (w/v) solid loading. The initial OD was adjusted to 10, and the fermentation was conducted in a 50 mL flask with a working volume of 10 mL. The process was carried out at 30°C and 250 rpm (Fig. 1B). All experiments were performed in biological triplicates.
To quantify metabolic products, including XOS, xylose, and glucose, fermentation products were collected at 24-h intervals. Collected samples were centrifuged at 20,000×g for 10 min. The supernatant was filtered through a 0.22 μm PES filter and analyzed using a high-performance liquid chromatography (HPLC) device equipped with a refractive index (RI) detector (Agilent Technologies) and a Shodex KS-802 column (Resonac). The column was eluted with HPLC-grade water at a flow rate of 0.5 mL/min at 80°C, with an injection volume of 10 μL. Cell density was measured at 600 nm (OD600) using a UV/Vis spectrophotometer (Hangzhou Allsheng Instruments).
Cell counting was performed using a counting chamber (Marienfeld-Superior) under 200× magnification on an OLYMPUS BX43 microscope. Samples were diluted with sterile distilled water, and cells were manually enumerated. For each count, cells within five 0.2 × 0.2 mm squares (four quadrants and the center) were counted.
The dry cell weight (DCW) was determined based on the relationship between OD and cell concentration, where an OD of 1.0 at 600 nm corresponds to 3 × 107 cells/mL, which is equivalent to 0.26 g dry cell/L, as determined using a laboratory spectrophotometer.
The composition of neutral detergent fiber (NDF), acid detergent fiber (ADF), and lignin were analyzed for raw corn cobs and corn cobs pretreated with 2% NaOH. These analyses were conducted at the Institute of Agricultural Science, Chungnam National University, following standard Association of Official Analytical Chemists (AOAC) methods. All measurements were performed in duplicate. Hemicellulose content was calculated as the difference between NDF and ADF, and cellulose content was derived by subtracting lignin from ADF. The mass balance of the process was calculated based on the composition of cellulose, hemicellulose, and lignin in the corn cobs, as well as the products derived from these components.
All statistical analyses were performed using IBM SPSS Statistics software ver. 27 (IBM). Data were subjected to one-way analysis of variance (ANOVA) to evaluate differences among treatment groups. Tukey’s honestly significant difference (HSD) test was used for post-hoc comparisons at a significance level of p < 0.05.
RESULTS
Xylanase, an enzyme from the glycoside hydrolase (GH) family, is capable of cleaving the β-1,4-glycosidic bonds in the xylan backbone. Among these, endo-type GH11 enzymes specifically target xylan substrates rather than cellulose and can be used to produce various types of XOS. In this study, a cell surface display system was employed to immobilize the GH11 family endo-xylanase (TsaGH11) derived from T. saccharolyticum on the yeast cell surface, facilitating the efficient production of XOS and yeast protein [12,16].
To immobilize the enzyme on the yeast cell surface, the enzyme must first be transported through the endoplasmic reticulum and Golgi apparatus to the plasma membrane via a secretion protein. After being directed to the cell wall by a signal peptide, the enzyme requires an anchor protein to prevent its release into the extracellular space and ensure its retention on the cell surface (Fig. 2). Therefore, the expression of a signal peptide, target protein, and anchor protein is essential for effective enzyme immobilization on the cell surface. In this study, MFα1 (α-factor preproleader sequence), a widely used signal peptide, was utilized [17]. Additionally, six anchor proteins were tested to identify the optimal anchor protein for endo-xylanase expression [18,19].
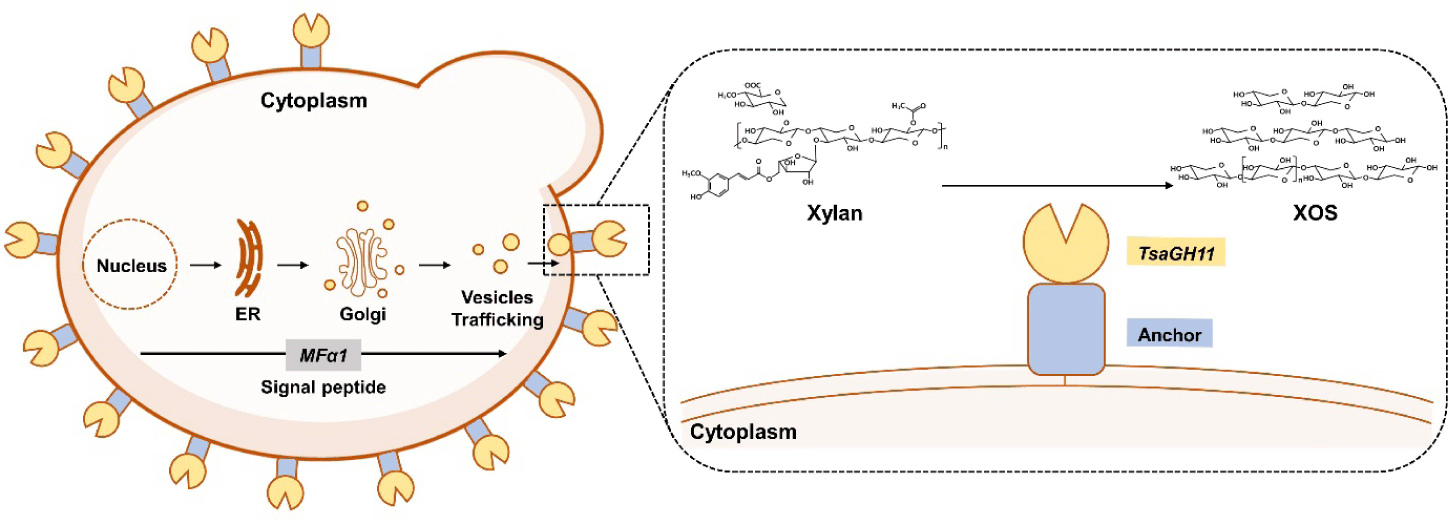
We constructed expression cassettes by fusing the endo-xylanase gene with the MFα1 signal peptide and six anchor proteins derived from S. boulardii. These cassettes were integrated into the intergenic region #1 of Chromosome VII in S. boulardii (Fig. 1A). As a result, six recombinant strains, designated X-Aga2, X-Cwp1, X-Cwp2, X-Sed1, X-Pir1, and X-Tir1, were constructed. Previous studies have demonstrated successful surface expression of eGFP in S. boulardii using the same approach [15].
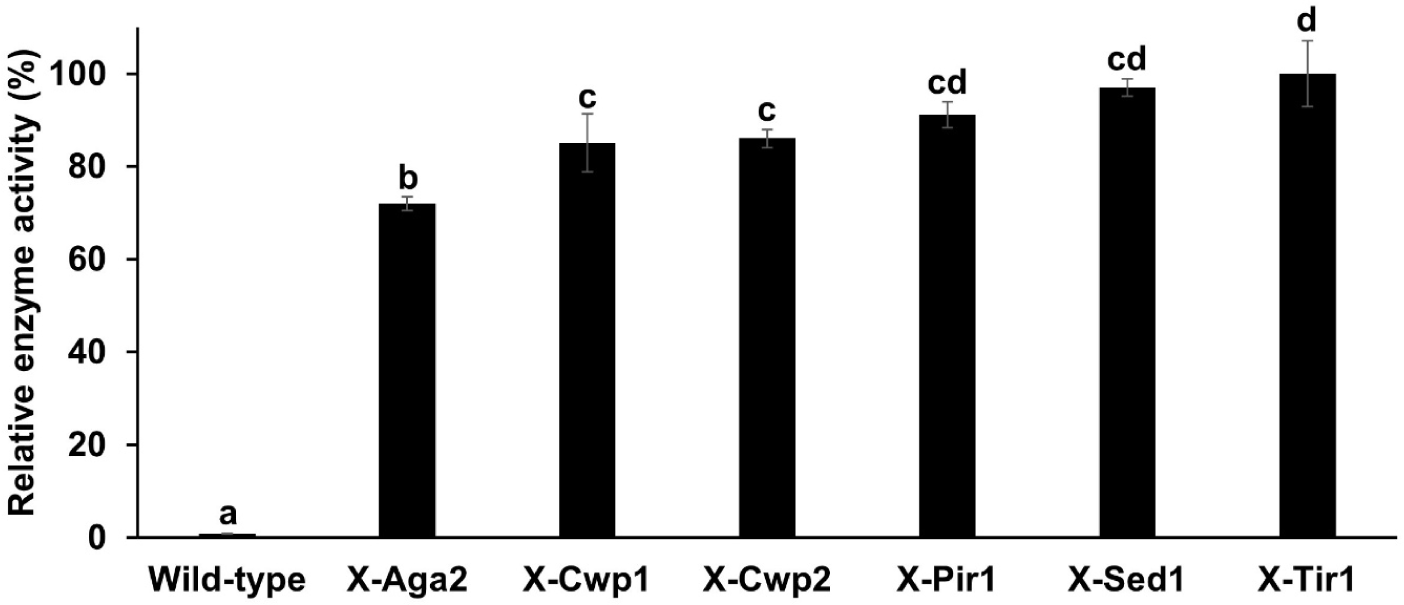
The enzyme activity of the six recombinant strains, each immobilized with a different anchor protein, was measured to compare expression efficiency. Since the yeast cells functioned as whole-cell biocatalysts, the strains themselves were directly used for enzyme activity assays. Using beechwood xylan as the substrate, all six recombinant strains exhibited higher enzyme activity than the wild-type S. boulardii after a 30-min reaction, indicating successful surface immobilization of endo-xylanase. Among the strains, X-Tir1 exhibited the highest enzyme activity and was selected for subsequent fermentation experiments (Fig. 3).
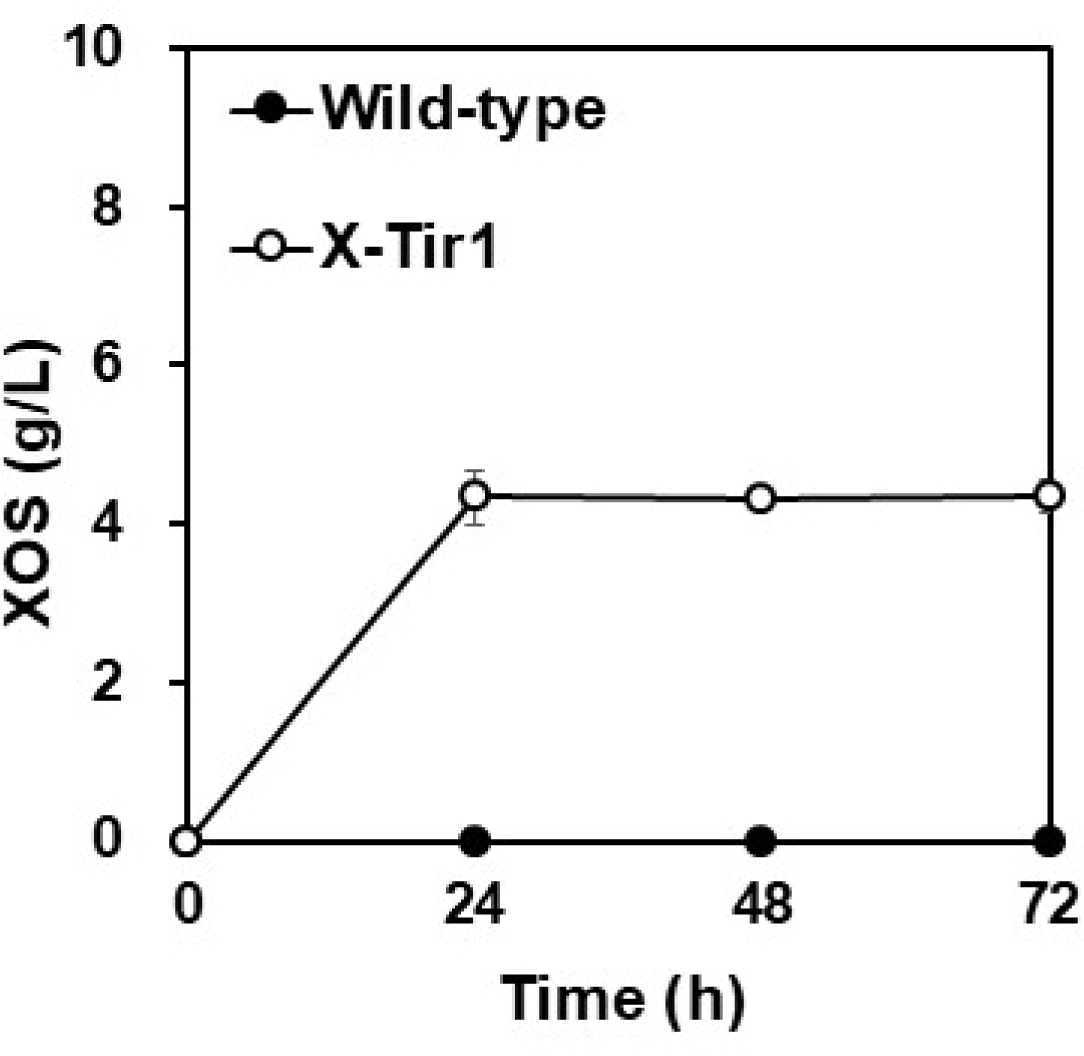
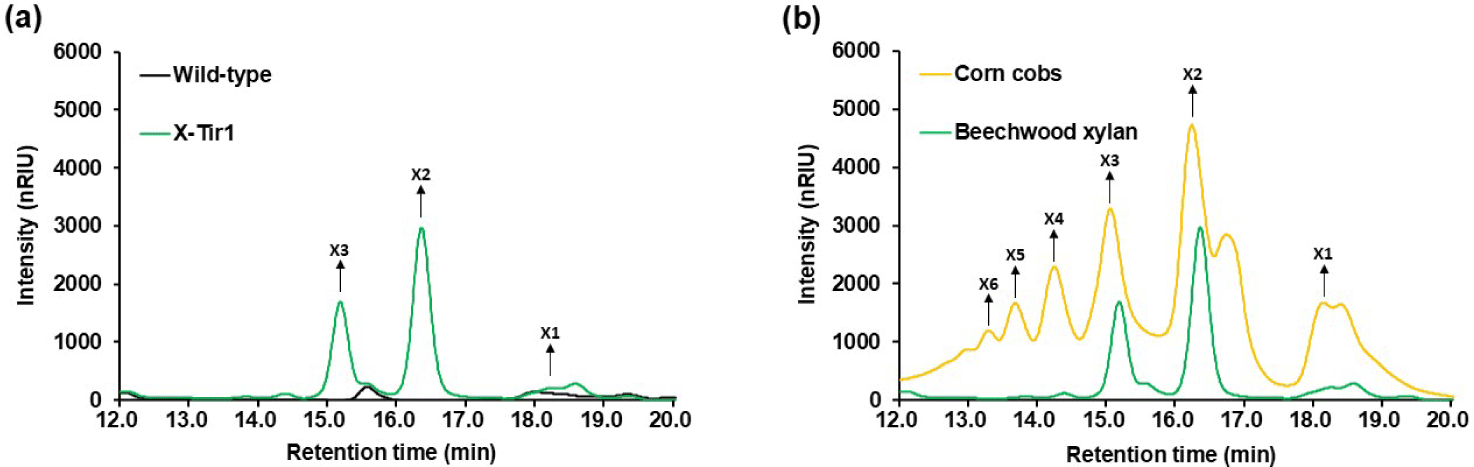
Prior to biomass fermentation, we confirmed that the selected X-Tir1 strain retained consistent enzyme activity during fermentation. Using 20 g/L of beechwood xylan as the substrate to test fermentation efficiency, the wild-type S. boulardii failed to degrade xylan. In contrast, the recombinant X-Tir1 strain produced approximately 4.3 g/L of XOS (X6–X2) after 72 h of fermentation (Fig. 4). When beechwood xylan was used as the substrate, peaks corresponding to lower degrees of polymerization (DP) XOS, such as X3 and X2, were primarily observed (Fig. 5A), which aligns with findings from previous studies [16]. This result confirms that the X-Tir1 strain maintained sufficient endo-xylanase activity under the mild conditions of 30°C and pH 6.5.
To efficiently utilize xylan from corn cobs, optimal pretreatment conditions were investigated. Among various biomass pretreatment methods, 1% H₂SO₄ pretreatment is one of the most widely used approaches [20]. In this study, solids treated with 1% H₂SO₄ produced approximately 3.4 g/L of XOS after 24 h of fermentation, likely attributed to the extensive hydrolysis of xylan into xylose. This observation is consistent with previous reports indicating that acid pretreatment primarily decomposes xylan into monosaccharides such as xylose [21]. The presence of approximately 9.4 g/L of xylose at the beginning of the fermentation process further supports this interpretation (Fig. 6B). Similarly, the heat-treated control group, which only received distilled water, exhibited low XOS production (Fig. 6A), likely due to insufficient breakdown of hydrogen and ester bonds between the components [22]. These results indicate that neither water nor acid pretreatment is suitable for efficient XOS production, highlighting the need for alkaline pretreatment.
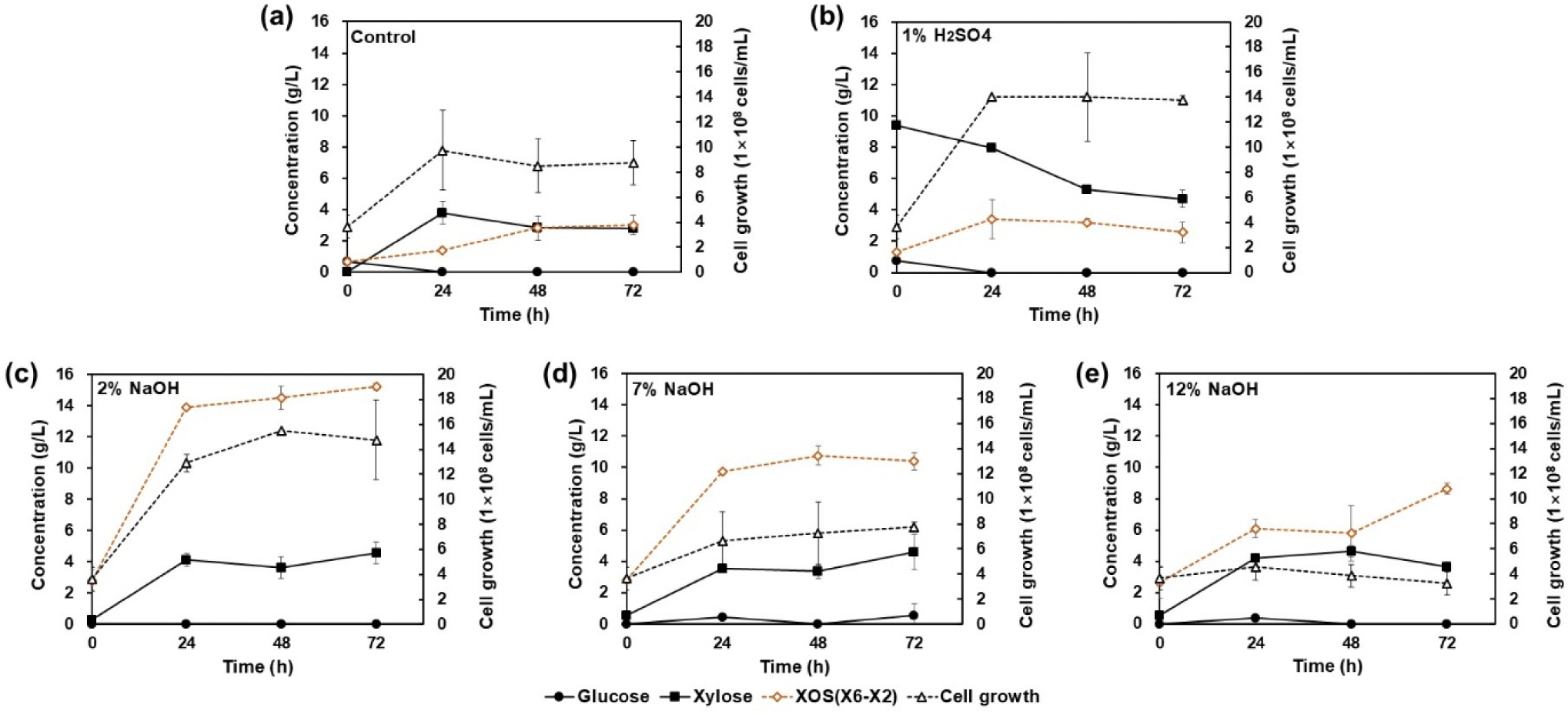
In contrast, alkaline pretreatment effectively breaks the hydrogen bonds between lignin-cellulose and hemicellulose-cellulose, as well as the ester bonds between lignin and hemicellulose, allowing for more efficient solubilization of xylan [23]. To identify the optimal alkaline pretreatment conditions, pretreatment was performed using 2%, 7%, and 12% (w/v) NaOH solutions and the results were analyzed. The solids pretreated with 2% (w/v) NaOH yielded the highest XOS production, approximately 15.2 g/L, after 72 h of fermentation (Fig. 6C). However, XOS production decreased with higher NaOH concentrations (Figs. 6C, 6D, and 6E), likely due to excessive solubilization of xylan into the liquid fraction during pretreatment, as well as increased salt formation during neutralization, which reduced the purity of xylan [23].
Furthermore, in contrast to fermentation using beechwood xylan as the substrate, fermentation with corn cobs produced XOS with varying DP, ranging from X2 to X6. This variation is likely attributed to structural differences in the xylan (Fig. 5B). Beechwood xylan has a simple glucuronoxylan structure, whereas corn cob xylan possesses a more complex glucuronoarabinoxylan structure with arabinose side chains [24,25].
Cell growth, which serves as an indicator of yeast protein production, was highest under the 2% NaOH pretreatment condition. In contrast, pretreatment with higher NaOH concentrations resulted in a significant decline in cell growth rates. This reduction is likely due to the inhibitory effects of salt formation during pretreatment [26]. Consequently, the 2% (w/v) NaOH pretreatment condition was determined to be optimal, yielding 15.2 g/L of XOS and 1.48 × 10⁹ cells/mL of yeast after 72 h of fermentation (Fig. 6).
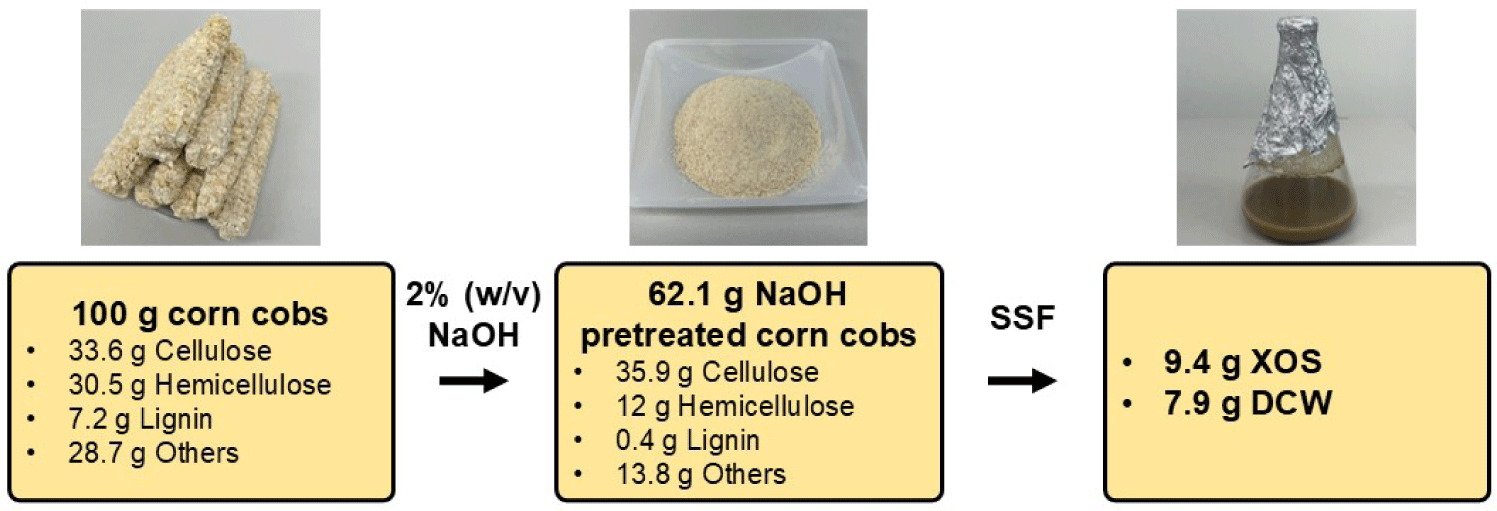
The initial 100 g of raw corn cobs contained 33.6 g of cellulose, 30.5 g of hemicellulose, 7.2 g of lignin, and 28.7 g of other components. Following pretreatment with 2% (w/v) NaOH, 62.1 g of pretreated corn cob solids were obtained, consisting of 35.9 g of cellulose, 12 g of hemicellulose, 0.4 g of lignin, and 13.8 g of other components.
Alkaline pretreatment effectively reduced the lignin content, which, in turn, enhanced the accessibility of cellulose and hemicellulose for enzymatic hydrolysis. Following SSF, 9.4 g of XOS and 7.9 g of DCW were produced from 62.1 g of pretreated corn cob solids (Fig. 7).
DISCUSSION
Over the past two decades, the market for XOS as a prebiotic has steadily expanded, prompting the development of various strategies to improve efficiency and maximize the yield of XOS production [27]. XOS can be produced from lignocellulosic biomass by using xylan, using production methods including chemical synthesis, enzymatic hydrolysis, or a combination of chemical and enzymatic treatments [12]. Among these, enzymatic hydrolysis offers several advantages over other methods, including control over the DP and the ability to produce functional XOS with high purity and low DP. Numerous studies have reported successful XOS production through enzymatic hydrolysis [28–30]. However, the extraction of xylan and enzyme purification remain critical steps, adding complexity to the process.
This study proposes the use of a cell surface display system to immobilize enzymes, which addresses some of these challenges. Enzymes immobilized on the cell surface can be easily reused through simple cell recovery, eliminating the need for a separate enzyme purification process. Additionally, they retain activity under mild reaction conditions, thereby enhancing reaction efficiency [19]. Recent research on cell surface display systems has emphasized the crucial role of anchor proteins in optimizing display efficiency [31]. In this study, we compared various anchor proteins, including Aga2 (Agglutinin type), Cwp1, Cwp2, Sed1, Pir1, and Tir1 (GPI type), as well as Pir1 (PIR type) [19]. A previous study utilized six different anchor proteins to express eGFP protein, with Sed1 demonstrating the most effective protein immobilization on the cell surface [15]. However, in this study, Tir1 exhibited the highest enzyme expression efficiency, suggesting that the optimal choice of anchor protein may vary depending on the target protein. Therefore, selecting the optimal anchor protein that aligns with the characteristics of the expressed protein and the specific substrate requirements is crucial for maximizing enzyme activity [32].
Research on cell surface display systems using S. boulardii has been limited, and this study demonstrates the potential to expand the application of display technology by utilizing S. boulardii strains. Recognized as Generally Recognized as Safe (GRAS), S. boulardii can be safely used in animal feed. This study employed CRISPR/Cas9-based genome integration instead of plasmid insertion, eliminating the need for antibiotic resistance genes, thus resulting in a safer strain for feed applications [33,34]. The strain developed in this study offers an innovative strategy for simultaneously supplying XOS and yeast protein through enzyme immobilization on the yeast cell surface, offering a safe solution for animal feed.
Corn cob has the potential to serve as an alternative fiber-based feed ingredient for livestock. However, its low palatability and high lignin content result in reduced nutritional value, poor digestibility, and limited feed efficiency. To overcome these limitations and enhance its nutritional value, microbial fermentation has been proposed as an effective strategy [35–37]. Previous studies have demonstrated that dietary supplementation with XOS significantly improves average daily gain (ADG), thereby enhancing growth performance and feed efficiency [38,39]. Additionally, yeast supplementation has been shown to improve feed efficiency, suppress intestinal inflammation, protect gut barrier function, and prevent E. coli infections, ultimately promoting gut health [40]. Although this study did not directly assess the functional effects of the fermented product as a feed additive, the findings from previous studies suggest that it may offer similar benefits. Future investigation through in vivo animal trials is warranted to validate its functionality and optimize its application.
The pretreatment process to enhance xylan accessibility was optimized. When 2%, 7%, and 12% (w/v) NaOH were applied to corn cob powder, high alkaline concentrations negatively impacted XOS production. Typically, high-concentration NaOH treatments solubilize glucan and xylan into the liquid fraction, reducing the amount of xylan remaining in the solid fraction and generating substantial amounts of salt (NaCl) during neutralization, which diminished the purity of xylan [23,41]. To address the issue of salt formation, NaOH removal steps, such as dialysis or membrane filtration, are required [23]. However, high-concentration alkaline pretreatment tends to reduce both solid recovery and XOS production, limiting its practical application. Therefore, for efficient utilization of xylan in the solid fraction, low-concentration alkaline pretreatment, such as 2% NaOH, is required [7]. In this study, 2% (w/v) NaOH pretreatment, which induced moderate delignification, was considered the optimal condition for enhancing xylan accessibility. Future studies should focus on utilizing the solubilized xylan from the pretreatment process to produce additional XOS or on valorizing lignin to generate high-value compounds such as furfural, vanillin, and syringaldehyde [7].
Fermentation of pretreated corn cobs produced XOS with varying DP from X2 to X6. The biological functions of XOS are closely linked to their DP, with beneficial gut bacteria such as Bifidobacterium and Lactobacillus typically favoring low DP X2 and X3 oligosaccharides [9,42]. Therefore, further studies are needed to explore the potential of enhancing the utilization of low-DP oligosaccharides by improving enzymatic hydrolysis efficiency, particularly through the addition of GH10 family endo-xylanase [24,30].
In this study, SSF was employed to simultaneously produce XOS and yeast protein from corn cobs. SSF integrates enzymatic hydrolysis and fermentation into a single process, simplifying the overall procedure. However, a general limitation exists, as the optimal temperature and pH for enzymatic hydrolysis and fermentation differ [43]. In this study, this limitation was addressed by immobilizing enzymes on the yeast cell surface, thereby simplifying the SSF process. In previous methods, XOS production was followed by treating residual solids with cellulase to convert them into fermentable monosaccharides, subsequently producing yeast protein [13,44,45]. In contrast, this study integrated these steps into a single SSF process, demonstrating the feasibility of simultaneously producing both XOS and yeast protein. While further optimization and scale-up studies are required, this approach offers a promising strategy for utilizing agricultural residues to develop functional feed additives in a more cost-effective and environmentally sustainable manner.
