INTRODUCTION
Short-chain fatty acids (SCFAs) are metabolic products produced during the microbial fermentation of non-digestible fibers [1–3]. The fermentation of amino acids also contributes to the production of SCFAs [3,4]. They are produced at high levels by bacteria in the proximal colon, where they are consumed by enterocytes [5]. The major SCFAs produced in the gut are formate, acetate, butyrate, and propionate, while lactate and other organic acids are produced as well [1,3]. The production of SCFAs involves a complex enzymatic process, commonly involving glycolysis, or, in other bacteria, pentose phosphate pathway [5]. Certain species of gut bacteria also facilitates metabolic cross-feeding, wherein, an SCFA (such as acetate or lactate) is converted into another SCFA (such as butyrate) [5,6]. SCFAs play an important role in metabolic homeostasis and overall gut health in humans and animals. These metabolites are known to improve gut integrity, regulate glucose and lipid homeostasis, and improve immune function [1,2].
Host diets play a crucial role in determining the level of SCFAs produced by gut microbiota [7,8]. Diets rich in fibers or supplemented by prebiotics can trigger the production of high levels of SCFAs [7,9,10]. In addition, previous studies have shown the ability of probiotics to modulate SCFA production, with regard to host health [11,12]. Dietary supplementation with a combination of microbial species, herein called multispecies probiotics (MSPs), was beneficial due to improved growth performance, reduction of incidence of diarrhea, and protection against pathogens in animals [13–16]. Although the modulatory effects of MSPs on host gut microbiota has been also examined in previous studies, there is still limited information on its effect on the modulation of gut microbiota and production of SCFAs in pigs, which is economically important in animal industry. Therefore, the goal of this study is to investigate the effects of MSP supplementation on the gut microbiota and fecal SCFA and lactate levels of weaned pigs. We also aim to examine the relationship between the gut microbiota and the levels of fecal SCFA after MSP feeding.
MATERIALS AND METHODS
A total of 38 pigs (Landrace × Yorkshire × Duroc) were used in this study, which were weaned at 4 weeks of age and had an average initial body weight of 23.53 ± 0.12 kg. The pigs were randomly divided into two groups: 18 pigs were placed in the control (CON) group, and the other 20 pigs in the MSP group. Each pen contained five pigs and was equipped with a one-sided self-feeder and nipple waterer for ad libitum access to feed and water. For 6 weeks, the control group continuously received the basal diet (Supplementary Table S1) without the administration of antibiotics or probiotics, while the probiotic (MSP) group received the basal diet supplemented with MSPs containing Bacillus amyloliquefaciens G10 (National Center for Biotechnology Information [NCBI] accession number CP072612), Levilactobacillus brevis M10 (NCBI accession number CP079699), and Limosilactobacillus reuteri RTR (NCBI accession number CP073062). These microbial species selected for the MSP supplement were isolated in a previous study for their potential probiotic characteristics in vitro (unpublished data). Individual probiotics were prepared using solid-state fermentation process, then processed into powdered form. The powdered probiotics were mixed with the basal feed to achieved their individual dosages. These processes were performed in the Genebiotech (Seoul, Korea). The dosages of B. amyloliquefaciens G10, L. brevis M10, and L. reuteri RTR added to the feed of the MSP group were 2.8×108 CFU/g feed, 2.3×107 CFU/g feed, and 4.8×106 CFU/g feed, respectively, and the viable cell counts were confirmed on the day of feeding. Fresh fecal samples were collected from the rectum of each pig before and after treatment, then stored at −20°C until further analyses. All animal protocols were approved by the Dankook University Animal Care Committee.
Fecal samples were prepared for high-performance liquid chromatography (HPLC) analyses as previously described [17]. The concentrations of SCFAs (formate, acetate, propionate, and butyrate) and lactate in pig fecal samples were determined via HPLC following the method described by De Baere et al. [18] with modifications. Briefly, 10 mM potassium dihydrogen phosphate (pH 2.2) and methanol (80:20 ratio) were used as the mobile phase with a C-18 reversed phase (150 × 4.6 mm, 5 µm) column (Young Jin Biochrom, Seongnam, Korea). HPLC was performed using a 1260 Infinity HPLC machine (Agilent, Santa Clara, CA, USA) equipped with a quaternary pump set, autosampler, column thermostat set at 35°C, and a multi-wavelength detector set at 210 nm.
Genomic DNA was extracted from the fecal samples using a QiaAmp PowerFecal DNA Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The concentration and purity of the genomic DNA were checked using a UV spectrophotometer (Mecasys, Daejeon, Korea). Amplification and sequencing of the V3–V4 hypervariable region of the 16S rRNA gene was performed using an Illumina MiSeq platform (Illumina, San Diego, CA, USA).
We performed alpha-diversity and taxonomic analyses of the raw paired-end sequences using EZBioCloud pipeline [19]. Then the samples were grouped into microbiome taxonomic profile sets for further analyses. Relative abundance cut-offs at the phylum, family, and genus levels were set to 0.1%. Charts depicting the results from the alpha-diversity and taxonomic analyses were generated using GraphPad Prism software version 8.4.2 (GraphPad, San Diego, California, USA). Principal Coordinate Analyses (PCoAs) were performed using Quantitative Insights Into Microbial Ecology software (QIIME, v. 1.9.0) [20] to calculate the β-diversity based on the Bray-Curtis distance metric, as previously described [21].
To illustrate the shared and unique microbiota found in each dietary group, a Venn diagram was created using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). To identify taxonomic markers for each group, linear discriminant analysis effect size (LEfSe) [22] analyses were performed using an online tool published by the Huttenhower Lab (https://huttenhower.sph.harvard.edu/galaxy/). To illustrate the correlation between the gut microbiota and fecal SCFAs and lactate contents, a heatmap was created using R (v.4.0.2) [23]. A co-occurrence network was illustrated using Cytoscape v.3.8 [24]. Predictions of the Kyoto encyclopedia of genes and genomes (KEGG) functional pathways (level 3) were performed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software [25]. Differential KEGG pathways between groups were identified using STAMP v.2.1.3 [26].
GraphPad Prism software version 8.4.2 (GraphPad) was utilized for statistical analyses. Data normality was analyzed using Shapiro-Wilk test. Significant differences were calculated using Kruskal-Wallis H test or Mann-Whitney U test, when appropriate. Permutational multivariate analysis of variance (PERMANOVA) was used to determine significant differences in PCoAs. Spearman’s rank correlation coefficients were calculated in R (v.4.0.2). For all statistical analyses, significance was determined at p < 0.05, p < 0.01, and p < 0.001.
RESULTS
The growth performance of the pigs after 6 weeks of feeding with MSP supplementation is summarized in Table 1. We found no significant differences in the body weight and average daily gain (ADG) between pigs in the MSP and CON groups (p > 0.05), indicating that MSP administration did not affect growth performance. On the other hand, the fecal concentrations of formate (1.4-fold; p < 0.05), acetate (1.8-fold; p < 0.05), and lactate (2.3-fold; p < 0.01) were significantly higher in the MSP group (Table 2), whereas propionate and butyrate were tended to increase by 12% and 14%, respectively with MSP feeding, but not significantly so (Table 2). These results indicate that MSP induced the significant increase in the production of lactate, acetate and formate directly and/or indirectly.
| Items | CON | MSP | p-values1) |
|---|---|---|---|
| Initial body weight (kg) | 23.61 ± 0.28 | 23.48 ± 0.26 | 0.74 |
| Final body weight (kg) | 48.66 ± 1.28 | 46.61 ± 1.30 | 0.20 |
| Average daily gain (ADG) (kg) | 0.60 ± 0.03 | 0.55 ± 0.03 | 0.27 |
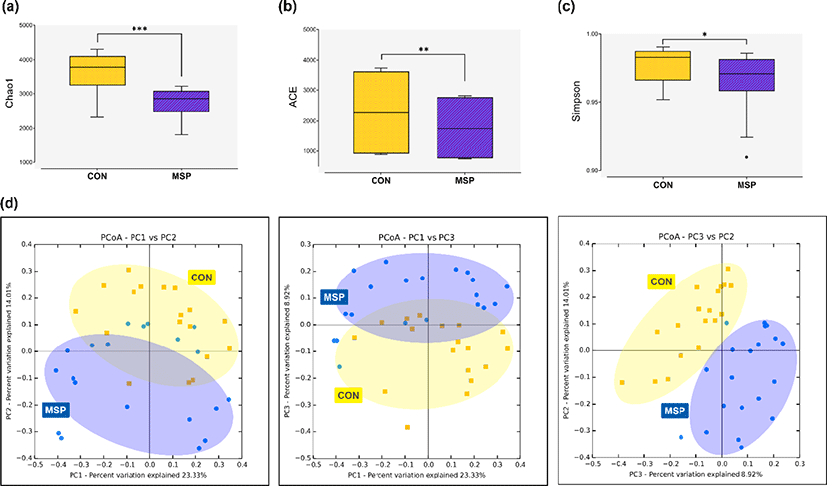
A total of 3,731,423 16S rRNA gene sequences were generated from the CON and MSP fecal samples. After removal of low-quality, nontargeted and chimeric amplicons, an average of 59,185 valid reads were generated. The sequencing depth was deemed sufficient for metagenomic analyses according to Good’s coverage scores for both the CON and MSP groups (98.7% and 99.4%, respectively). The alpha diversity indices Chao1 (p < 0.001) and abundance-based coverage estimator (ACE, p < 0.01) were significantly lower in the fecal samples of the MSP group (Figs. 1a and b). By contrast, the Simpson index was significantly higher in MSP samples (p < 0.05) (Fig. 1c). Moreover, the PCoA plot constructed based on Bray-Curtis dissimilarity index revealed that the MSP fecal microbiota clustered separately from that of the CON group (Fig. 1d; PERMANOVA, p < 0.001). Our results strongly suggest that MSP supplementation altered the gut microbiota structure of the pigs.
At the phylum level, we observed five abundant phyla among the groups (abundance cut-off was set at 0.1%; Fig. 2a; Supplementary Table S2). Firmicutes and Bacteroidetes were the most abundant phyla across the groups, covering about 90% of the total sequence reads. We found that MSP supplementation significantly increased the abundance of Firmicutes (59.07% vs. 65.67% for CON vs. MSP, respectively; p < 0.01), and decreased the abundance of Bacteroidetes (34.27% vs. 24.41% for CON vs. MSP, respectively; p < 0.01). Consequently, the Firmicutes:Bacteroidetes (F:B) ratio was significantly higher in the MSP group compared to in the CON group (1.72 vs. 2.69 for CON and MSP, respectively; p < 0.001). Moreover, the abundance of the phylum Actinobacteria was remarkably enriched following MSP supplementation (0.90% vs 5.61%; p < 0.01). By contrast, the abundance of the phylum Chlamydiae was greatly reduced after MSP supplementation (0.19% vs. 0.0003%; p < 0.05). At the family level, Ruminococcaceae, Prevotellaceae and Lachnospiraceae were the most abundant bacterial families (Fig. 2b; Supplementary Table S3). The intake of MSP lowered the abundance of Prevotellaceae (19.80% vs. 12.84%; p < 0.01).
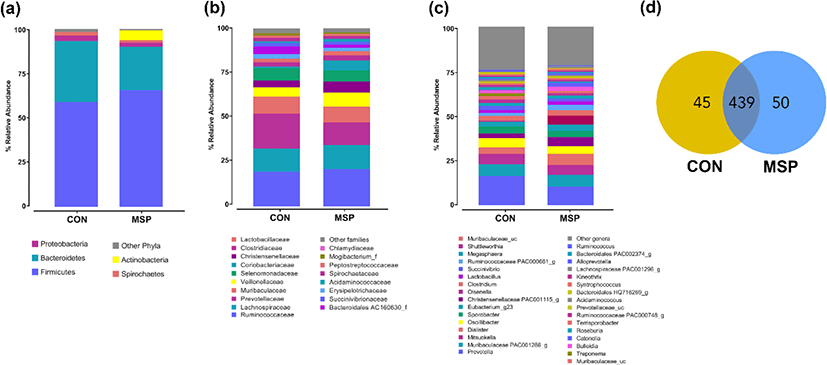
At the genus level, we identified a total of 534 genera across the groups (Fig. 2d), of which 439 were shared between the groups, while 45 and 50 were unique to the CON and MSP groups, respectively. Fig. 2c and Supplementary Table S4 summarizes the distribution of the genera between the groups. Prevotella was the most abundant genus in both groups. Supplementation with MSP led to the significant enrichment of Olsenella (0.06% vs. 5.20%; p < 0.01), Catonella (0.32% vs. 2.20%; p < 0.05), Acidaminococcus (0.29% vs. 1.19%; p < 0.01), Dialister (3.51% vs. 6.14%), Christensenellaceae PAC001115 (2.77% vs. 5.00%), Ruminococcaceae PAC000661 (1.58% vs. 2.81%; p < 0.05), and Catenibacterium (0.21% vs. 0.45%; p < 0.05). In addition, the abundance of Lactobacillus was also slightly higher (1.83% vs. 2.13%) in the MSP group. On the other hand, populations of Prevotella (16.40% vs. 10.45%; p < 0.01), Roseburia (1.51% vs. 0.67%; p < 0.05), Alloprevotella (1.03% vs. 0.52%; p < 0.01, Escherichia (0.32% vs. 0.05%), and Chlamydia (0.19% vs. 0.0003%; p < 0.05) were significantly lower in MSP-fed pigs.
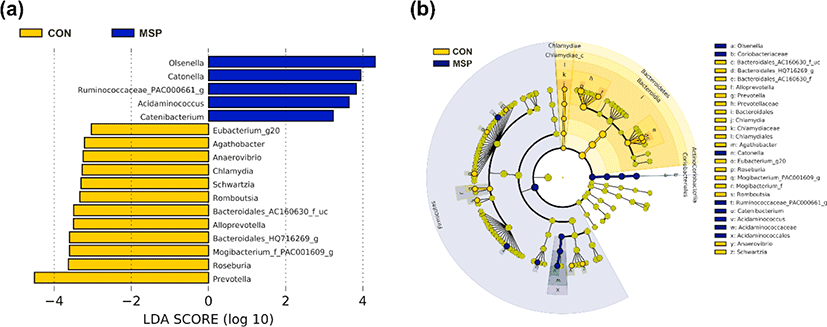
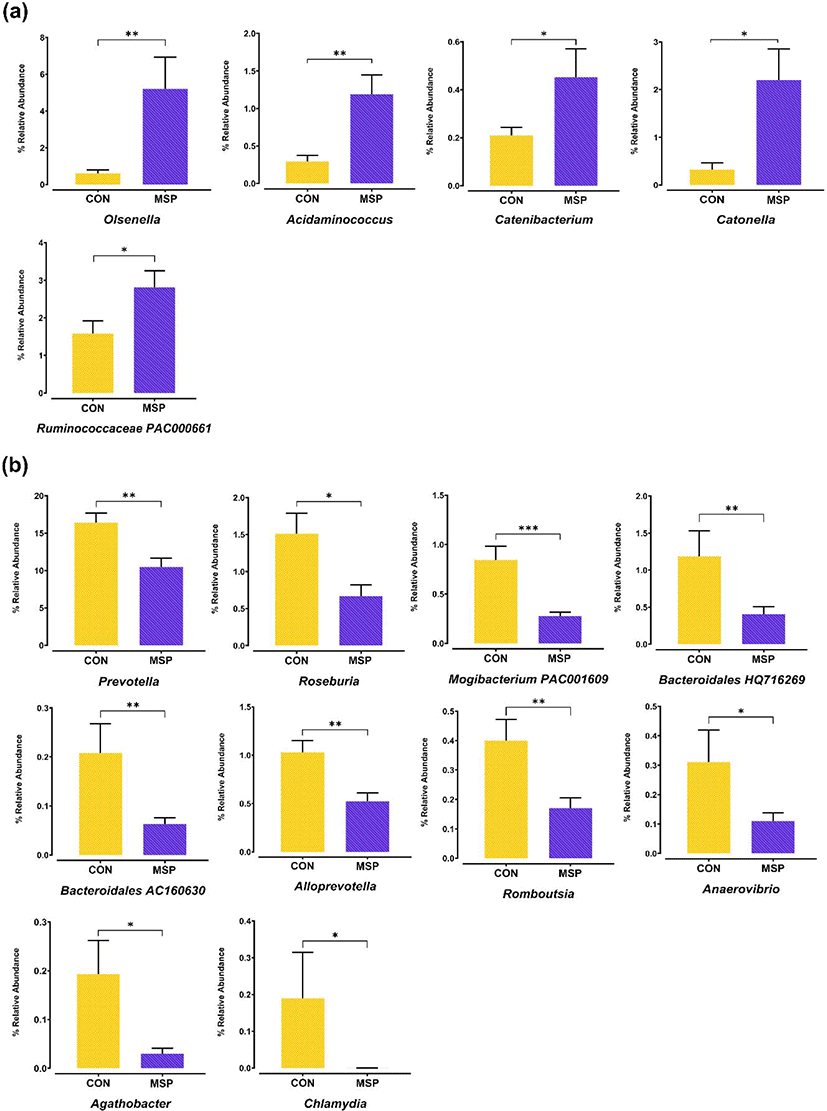
Next, we identified potential taxonomic markers in the gut microbiota of the pigs using LEfSe (Fig. 3). The enriched abundances of Olsenella, Catonella, Ruminococcaceae PAC000661, Acidaminococcus, and Catenibacterium were identified as taxonomic markers for the MSP group (p < 0.05). By contrast, Prevotella, members of the order Bacteroidales (AC160630, HQ716269), Mogibacterium PAC001609, Romboutsia, Anaerovibrio, Agathobacther, Alloprevotella, Roseburia, Schwartzia, and Eubacterium_g20 were identified as taxonomic features of the CON group (p < 0.05). Plots depicting the abundances of the taxonomic markers are shown in Fig. 4. Our results further suggest that MSP supplementation elicits a modulatory effect on the gut microbiota of pigs.
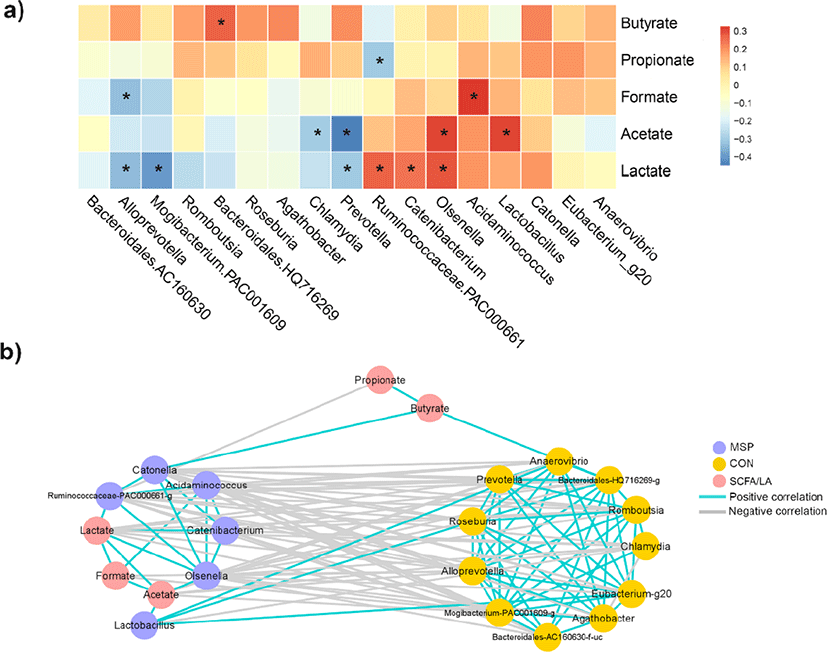
To identify the associations between the gut microbiota and the fecal SCFAs and lactate concentrations, we conducted a correlation analysis by calculating the Spearman’s correlation coefficients (Fig. 5a). It showed that Olsenella,Catenibacterium, and Ruminococcaceae PAC000661 have significantly correlated with lactate (p < 0.05). Olsenella and Lactobacillus were observed to be significantly associated with acetate (p < 0.05), while Acidaminococcus was significantly correlated with formate (p < 0.05). By contrast, Prevotella (p < 0.05), Mogibacterium (p < 0.01), and Alloprevotella (p <0.05) had significant negative correlations with lactate. In addition, Prevotella and Chlamydia were observed to be negatively linked with acetate (p < 0.05), while Alloprevotella was negatively associated with formate (p < 0.05). On the other hand, order Bacteroidales HQ716269 (p < 0.05), Roseburia, Agathobacter, Alloprevotella, Prevotella, and Anaerovibrio were positively linked with butyrate. These observed associations were supported by the network map (Fig. 5b), showing positive correlation among MSP-enriched taxa, lactate, formate, and acetate; Catonella has positive relationship with butyrate. Meanwhile, MSP-diminished taxa co-occurred with each other; order Bacteroidales HQ716269 positively correlated with butyrate. These observations suggest the potential interplay between the MSP-modulated gut microbiota and the production of SCFA and lactate.
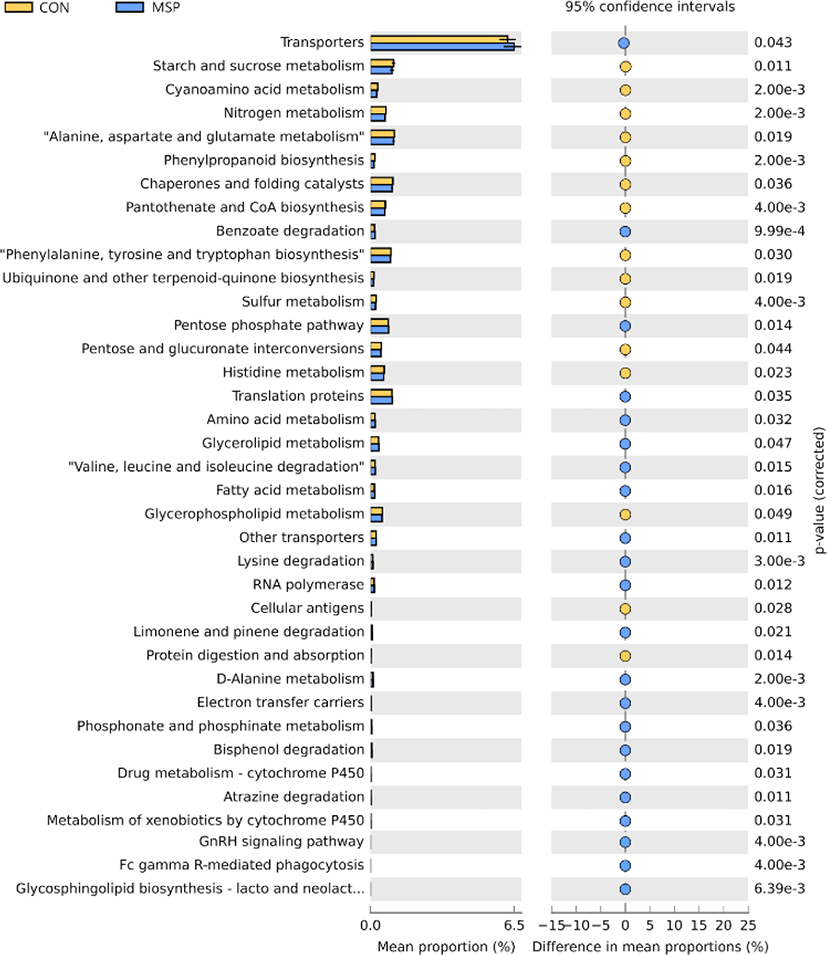
To predict functional markers in the gut microbiome of the pigs, we performed PICRUSt analyses [25]. A total of 267 KEGG pathways were predicted across both groups (Supplementary Table S5). We found that 30 of the predicted KEGG pathways were significantly altered in the MSP group (Fig. 6). KEGG pathways including transporters, pentose phosphate pathway, amino acid metabolism, glycerolipid metabolism, and fatty acid metabolism were significantly enriched in the MSP group compared to controls (p < 0.05). Other KEGG pathways, such as starch and sucrose metabolism, cyanoamino acid metabolism, nitrogen metabolism, alanine, aspartate and glutamate metabolism, phenylpropanoid biosynthesis, chaperones and folding catalysts, and pantothenate and CoA biosynthesis were enriched in the CON group (p < 0.05). This indicates that MSP supplementation altered the functional capacity of the pig microbiota.
DISCUSSION
We investigated the effects of supplementation with MSP containing B. amyloliquefaciens G10, L. reuteri RTR, and L. brevis M10 on the growth performance and fecal SCFA levels in weaned pigs. Our in vitro studies showed no antagonistic effect among these strains, but showed synergistic effect on the diversity of fecal microbiota in pigs (data not shown). Some previous studies have suggested that MSP supplementation in pigs leads to improved growth performance [13,14]. However, in the present study, no significant differences in body weight or ADG were observed in MSP-fed pigs. This observation is similar to those of other studies, in which no observable increase on body weight or ADG in pigs treated with probiotics was found [27–30]. This phenomenon may be due to multiple factors influencing body weight gain such as feeding concentration and period, environmental stress, animal age, or the specific characteristics of the probiotic strains [29,30].
Multiple studies have revealed the role of SCFAs in metabolic homeostasis and overall gut health in humans and animals. SCFAs (mainly acetate, butyrate, and propionate) are energy source for enterocytes, and known to improve gut integrity, regulate glucose and lipid homeostasis, and improve immune function [1,2,31]. Production of SCFAs and lactate by the gut microbiota has been extensively described in other studies [5,31]. The modulating effect of SCFAs on the host’s health is mainly attributed to its involvement in the inhibition of histone deacetylases and activation of G-protein-coupled receptors, such as GPR41, GPR43, and GPR109A [5,32]. Host diets play a crucial role in determining the level of SCFAs produced by gut microbiota, such that, diets rich in fibers or supplemented by prebiotics can trigger the production of high levels of SCFAs [7,9,10]. In addition, probiotics have demonstrated the ability to affect the production of SCFAs [11,12]. In the present study, we investigated the effect of MSP supplementation on the SCFAs and lactate levels in pigs. After the feeding period, we found a significant increase in the fecal levels of formate, acetate, and lactate in MSP-fed pigs. Similarly, Nagata et al. [33] and Alcon-Giner et al. [34] reported increases in acetate level following probiotic treatment. Acetate improves metabolic homeostasis, energy utilization, and appetite regulation [35,36]. The role of formate in gut health is not yet fully understood, although it can impart protective properties against pathogens such as Escherichia coli and Salmonella [37,38]. Lactate, which is mainly produced by lactic acid bacteria and bifidobacteria, serves as an intermediate molecule for acetate, butyrate, and propionate production by lactate-utilizing bacteria [4,39]. Although the fecal levels of butyrate and propionate were slightly higher in MSP-fed pigs, no significant differences were found in the present study, as observed previously in other studies [27,28].
At the phylum level, we observed elevated populations of Firmicutes in MSP-fed pigs compared to in the control group. A positive correlation between the abundance of Firmicutes and acetate and lactate levels has previously been reported [40,41]. Moreover, we found that the phylum Actinobacteria was also enriched following MSP supplementation. By contrast, MSP supplementation drastically reduced the population of the phylum Chlamydiae, which is responsible for various diseases in pigs [42].
At the genus level, we found that MSP supplementation caused the significant enrichment of Olsenella, Catonella, Acidaminococcus, Catenibacterium, and Ruminococcaceae PAC000661; these genera formed positive relationships with increased levels of acetate, formate, and lactate. Increases in the abundance of Olsenella have been observed in several studies that have examined the effects of supplementing diets with probiotics, complex carbohydrates, or prebiotics [43,44]. Olsenella is a carbohydrate fermenter and producer of acetate and lactate [45,46], which is consistent with our correlation results. Likewise, the genus Catenibacterium is positively associated with high SCFA levels [47]. Acidaminococcus can convert glutamate into acetate and butyrate [48] and Catonella enrichment is correlated with reduced diarrheal severity and an increase in acetate and butyrate levels in patients with irritable bowel syndrome treated with synbiotics [49]; this suggests that the prevalence of Acidaminococcus and Catonella improved the fecal level of acetate in MSP-fed pigs. In addition, the population of Lactobacillus was observed to increase upon MSP-feeding, and has shown to have positive correlation with acetate and lactate. Relative abundances of Bifidobacterium and other lactic acid bacteria was below cut-off (0.1%) for this study. The increase in Lactobacillus may be due to the supplementation of probiotics, which is similar to previous study [50]. Finally, Ruminococcaceae produces SCFAs from complex plant materials [51]. Elevated populations of Ruminococcaceae are negatively associated with constipation, inflammation, and liver diseases, and are typically observed in healthy subjects [52,53]. Our results indicate the potential positive effects of dietary MSP inclusion on pig health, especially on SCFA production, which should be investigated further.
MSP supplementation also decreased the abundance of Prevotella, Roseburia, Alloprevotella, Romboutsia, Anaerovibrio, Mogibacterium, the order Bacteroidales, Agathobacter, and Eubacterium, as revealed by LEfSe analyses. Furthermore, we found positive associations between order Bacteroidales, Prevotella, Roseburia, Agathobacter, and Alloprevotela, and butyrate levels. Our results are consistent with the fact that several members of the order Bacteroidales produce butyrate [54]. Prevotella has an increased prevalence in subjects with plant-rich diets and produces SCFAs [55,56]. Likewise, Alloprevotella and Anaerovibrio produce SCFAs [57–59]. However, elevated abundance of Prevotella are associated with inflammation, hypertension, insulin resistance, and liver disease [60,61]. In addition, Alloprevotella and Anerovibrio are present in individuals with colitis and chronic kidney disease [62,63], and rats with type 2 diabetes [64], respectively. Roseburia and Agathobacter are beneficial gut bacteria that produce SCFAs, particularly butyrate [65,66]. Our findings support these studies in that the abundance of these genera positively correlated with butyrate. Our results suggest that the decrease in the abundances of Prevotella, Roseburia, Agathobacter, the order Bacteroidales, Alloprevotela, and Anaerovibrio may have lowered the butyrate levels in the MSP-fed group due to a decrease in metabolite cross-feeding from acetate or lactate into butyrate [67,68]. Further studies are needed to investigate the possible interplay between probiotics and the fermentation capacity of these genera.
Interestingly, we observed a dramatic decline in the abundance of Chlamydia and Escherichia after supplementation with MSPs. Furthermore, we found that Chlamydia negatively correlated with acetate and lactate. Both Chlamydia and Escherichia are recognized pathogens in both humans and pigs [42]. SCFAs impart protective effects against enteric pathogens, either by inducing pH stress or by promoting inflammasomes [69,70]. Our results provide further evidence for enhanced SCFA production by MSP supplementation and their beneficial effects of gut health.
Finally, our PICRUSt analyses revealed that MSP supplementation also altered the metabolic functions of the gut microbiome. MSP treatment enriched several KEGG pathways including transporters, pentose phosphate pathway, amino acid metabolism, glycerolipid metabolism, and fatty acid metabolism. The elevated abundances of Firmicutes, as well as Lachnospiraceae and Ruminococcaceae may explain the abundance of transporters in MSP-fed pigs [5,51]. Moreover, the gut microbiota (mainly Firmicutes) utilizes the pentose phosphate pathway to degrade complex fibers, thereby producing SCFAs [71,72]. Amino acid metabolism by gut microbiota may also provide substrates for SCFA production [73]. Probiotic supplementation can increase the abundance of glycerolipid metabolism and fatty acid metabolism pathways, which are associated with improved host metabolic health [74,75]. These MSP-mediated changes in functional pathways further support our findings that MSP supplementation can modulate the gut microbiota and SCFA levels in pigs.
In conclusion, we found that MSP supplementation resulted in significantly higher levels of fecal acetate, formate, and lactate in pigs. Moreover, MSP supplementation altered the gut microbiota of the pigs by increasing the population of potentially beneficial bacteria such as Olsenella, Catonella, Catenibacterium, Acidaminococcus, and Ruminococcaceae. Dietary MSP also caused a decline in the abundance of pathogenic bacteria such as Escherichia and Chlamydia. The modulation of the gut microbiota was observed to be strongly correlated with the changes in fecal SCFAs and lactate levels, which suggests an interplay between the two. Furthermore, we found changes in the functional pathways present within the gut microbiome. Our results demonstrate the potential use of MSPs for improving the gut health of animals and humans. Further in-depth investigations are needed to explore the interplay between the taxa identified in this study and SCFAs and its overall effect on host metabolic health.
SUPPLEMENTARY MATERIALS
Supplementary materials are only available online from: https://doi.org/10.5187/jast.2021.e94.
















