ANNOUNCEMENT
Ligilactobacillus agilis, commonly isolated from animals, has been used as a representative species for the study of the motility of lactic acid bacteria, because it has motility features that are uncommon in lactobacilli [1,2]. Very recently, bacterial culture and cell-free supernatant of L. agilis 32 isolated from pig manure were reported to inhibit the growth of Enterotoxigenic Escherichiacoli 10 (ETEC 10), one of the causative agents of post-weaning diarrhea in piglets [3]. In this announcement, we report the genome of L. agilis C7, which inhibits Listeria monocytogenes (unpublished data), a frequent contaminant of many foods including dairy and meat products, and the three bacteriocin gene clusters encoding garvicin Q, salivaricin A, and a Blp family class II bacteriocin.
L. agilis C7, which showed antibacterial activity against L. monocytogenes ATCC 19114 in the agar-well diffusion assay (unpublished data), was isolated from piglet fecal samples. L. agilis C7 was cultured in de Man, Rogosa and Sharpe (MRS) broth and streaked on MRS agar. MRS agar plates were incubated at 37°C for 24 h. Genomic DNA was extracted from the C7 strain using the conventional phenol–chloroform protocol and was sent to Macrogen (Seoul, Korea) for whole-genome sequencing. The PacBio Sequel System (Pacific Biosciences, Menlo Park, CA, USA) and Illumina Platform (Illumina, San Diego, CA, USA) technologies were used to generate long and short sequence reads, respectively. PacBio sequencing generated a total of 2,235,708,853 bases with 237,414 total long reads (subreads), while Illumina sequencing generated a total of 800,163,663 bases and 5,301,328 paired-end reads after filtering. The Microbial Assembly Application was used for de novo assembly of the PacBio reads. Pilon (v1.21) was used to improve the initial assembly by mapping the Illumina reads, generating a more accurate assembly. The genome assembly of L. agilis C7 has a total size of 2,961,932 base pair (bp) with four contigs. Quality statistics were generated using QUAST (v5.0.2) and Benchmarking Universal Single-Copy Orthologs (BUSCO) (v5.3.2). QUAST reported the largest contig and N50 length of 1,727,998 bp and a guanine + cytosine (G + C) content of 40.15%, while BUSCO reported 124 complete and single-copy BUSCOs. Average Nucleotide Identity, ANIb option (pyani v0.2.10) identified L. agilis with > 95% sequence identity to publicly available L. agilis genomes.
Annotation of L. agilis C7 revealed 93 transfer RNAs, 24 ribosomal RNAs, 1 transfer-messenger RNA, 7 repeat regions, 2946 coding sequences, and 3116 genes (Table 1). BAGEL4 [4] identified two bacteriocin gene clusters encoding the bovicin 255 peptide variant and salivarin A, while AntiSMASH [5] mapped RiPP-like bacteriocin IIc (Fig. 1). The genome size of L. agilis C7 is relatively larger (~3.0 Mb) than those of other L. agilis strains (~2.0–2.5 Mb) in the NCBI database. Mobile genetic elements such as an integrative and conjugative element (~255 kb) and the 17 prophage regions (7 intact prophages) were also mapped to the genome of L. agilis C7 via VRprofile [6].
| Features | |
|---|---|
| Genome size (bp) | 2,961,932 |
| No. of contigs | 4 |
| N50 | 1,727,998 |
| GC content (%) | 40.15 |
| CDS | 2,946 |
| Genes | 3116 |
| tRNA | 93 |
| Misc RNA | 52 |
| rRNA | 24 |
| Repeat region | 7 |
| tmRNA | 1 |
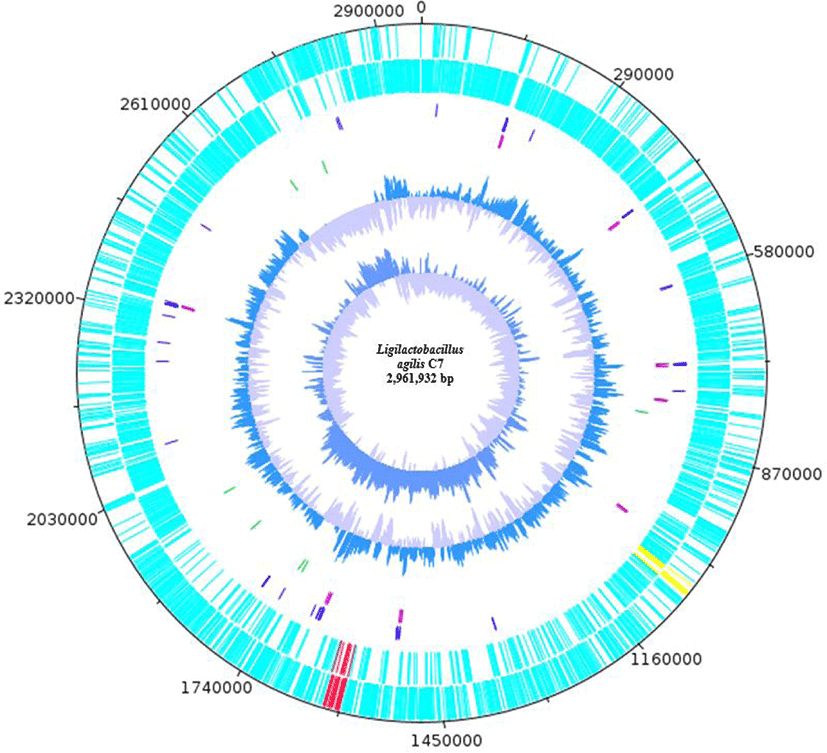
Blastp analyses of the bovicin 255 peptide variant, salivaricin A, and the RiPP-like bacteriocin IIc revealed 100% identity to garvicin Q family class II bacteriocin from other L. agilis strains, 62.5% identity to type A2 lanthipeptide from Streptococcus pyogenes, and 51.46% identity to Blp family class II bacteriocin from S. hyointestinalis, respectively. Garvicin Q is a subclass IId bacteriocin that was initially isolated from Lactococcus garvieae BCC 43578 and has been reported to be active against L. monocytogenes and several bacterial species belonging to the genera Bacillus, Enterococcus, Lactobacillus, Lactococcus, and Pediococcus [7]. Meanwhile, salivaricin A, which is active against Micrococcus luteus [8], is among the lantibiotics produced by strains of S. salivarius, although genetic variants have also been found in S. pyogenes [8,9]. Lantibiotics are ribosomally synthesized molecules that are heat-stable and reported to have therapeutic potential in treating infectious diseases. On the other hand, Blp family class II bacteriocin has been shown to mediate intraspecies competition among pneumococci [10].
This report highlights the potential of L. agilis C7 for the production of bacteriocin to control L. monocytogenes in dairy and meat products. In addition, this paper contributes to the scarce genetic information on L. agilis.
















