INTRODUCTION
The small intestine consists of the duodenum, jejunum, and ileum and performs a variety of functions, such as nutrient absorption, electrolyte uptake, hormone secretion and host-pathogen interactions in intestinal epithelium that is composed of a variety of intestinal cell types such as Paneth cells, enteroendocrine cells, goblet cells and enterocytes [1,2]. Recently, a scaffold-based three-dimensional (3D) culture system has provided a reliable alternative platform for the establishment of pluripotent and adult stem cells derived intestinal organoids in vitro. in vitro 3D organoid systems are valuable tools and potential alternatives to in vitro systems [3], and these in vitro organoid systems have been used for novel practical applications in various fields, such as animal science, animal biotechnology, and biomedicine [1].
Recent progress has been made to establish intestinal organoids derived from livestock, including bovine [2], porcine [4], chicken [5] and equine [6]. In particular, we previously reported a reliable method for the isolation of intestinal crypts from small intestines and the robust 3D expansion of intestinal organoids (basal-out) derived from adult bovine [2,7]. Genearally, basal-out intestinal oragnoids have the intestianl basal membrane on the surface. However, there are practical limitations associated with the potential use of intestinal organoids, such as host-pathogen interactions and nutrient absorption, because intestinal organoids cultivated in Matrigel have all basal-out structures [8–10]. Thus, development of apical-out organoid culture system (polarity reversal) is required. Moreover, very few studies on the functional characterization and 3D expansion of adult stem cells isolated from livestock species compared to those from other species are available [11].
In this study, we successfully established growing cattle-derived intestinal organoids and apical-out intestinal organoids and characterized their cellular potentials using gene expression profiling and immunocytochemistry. Furthermore, we investigated the permeability of the epithelial barriers of these apical-out and basal-out organoids using fluorescein isothiocyanate (FITC)-dextran.
MATERIALS AND METHODS
The aim of present study was the identification of adult intestinal stem cells and the cultivation of intestinal organoids derived from growing cattle (12 months old). Hanwoo growing cattle (Bos Taurus coreanae) were used for these experiments, and the experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the National Institute of Animal Science (NIAS-2019-366), Korea.
The isolation of intestinal crypts from growing cattle and establishment of intestinal organoids were conducted as previously described [2]. Briefly, ileum and jejunum fragments were obtained from the small intestines of growing cattle (12 months old). After washing ice-cold phosphate-buffered saline (PBS) and 1% penicillin/streptomycin (Sigma-Aldrich, New York, NY, USA), The fragments were dissected vertically and washed thoroughly with washing buffer to remove debris. To remove the mucosal and submucosal layers were used to scrape a glass slide. The remaining muscle layer was cut into 3- to 5-mm pieces and repeatedly washed by shaking vigorously until the supernatant was clear. After centrifuged at 300×g, the collected pellet was resuspended in 25 mL of Cell Disassociation Solution (Stem Cell Technologies, Vancouver, Canada) and incubated at room temperature for 40 min on a rocker to release the crypts. Intestinal crypts were added in 1 mL of intestinal human organoid medium (Stem Cell Technologies) and the number of intestinal crypts was counted using a hemocytometer under an inverted microscope. A total seeding mixture with 100–150 crypts and Matrigel in a 1:1 ratio was prepared and carefully placed in the middle of the wells of a 24-well plate. After 20 min, the matrigel dome was completely polymerized and gently added 1ml of organoid growth medium to each well.
The growing cattle-derived intestinal organoids were passaged approximately once a week upon maturation as previously described [2]. Briefly, the medium was completely removed and the organoid dome was rinsed with ice-cold PBS. The organoids were collected by centrifugation at 300×g for 5 min after removing the Matrigel dome using enzyme-free cell disassociation buffer. The pellets were seeded in the amount of medium and Matrigel in a 1:1 ratio, and each well (140–150 organoids) was passaged into three parts in 24-well plates. The medium was replaced every 3 days, and the organoids were subcultured once a week. The number of organoids was counted in triplicate every week. For cryopreservation, the organoids were resuspended in preserving solution composed of 90% medium and 10% dimethyl sulfoxide (Sigma-Aldrich), and transferred to a liquid nitrogen tank after stored at −80°C for 24 hr.
After being thoroughly washed with ice-cold PBS, ileum and jejunum fragments from the small intestine were fixed in 10% neutral-buffered formalin (Sigma-Aldrich). The fragments were subsequently embedded in a paraffin block, and the fragments were vertically and horizontally sectioned at a thickness of 3–5 μm. The sections were then deparaffinized in xylene, rehydrated with water using a graded series of alcohol solutions, and processed prior to hematoxylin and eosin (Merck, Darmstadt, Germany) staining. For immunohistochemical analysis, the sections were permeabilized with 0.1% Triton X-100 in PBS for 5 min and incubated with 0.1% normal goat serum for 1 h to block nonspecific binding after antigen retrieval by boiling in a sodium citrate buffer solution. The samples were incubated overnight at 4°C with the appropriate primary antibodies. The antibodies used in the present study are shown in Table 1. After washing with PBS, the samples were incubated with anti-mouse and anti-rabbit secondary antibodies coupled to Alexa Fluor-488 and Alexa Fluor-594 (Molecular Probes, Eugene, OR, USA), for 1 h at room temperature, respectively. Diamidino-2-phenylindole (DAPI) was used to counterstained these fluorescent samples. Images were captured using an Olympus X100 confocal microscope (Olympus, Tokyo, Japan).
The organoids were maintained in 24-well plates until maturation. The medium was removed from the wells, and the organoids were gently washed by cold PBS. Briefly, the organoids were incubated in neutrally buffered 4% paraformaldehyde (Sigma-Aldrich) for 30 min at room temperature. After permeabilization with 0.5% Triton X-100 (v/v) (Sigma-Aldrich) in PBS for 30 min at room temperature, the blocking step was performed using 3% bovine serum albumin in PBS for 1 h at room temperature. Then, the organoids were thoroughly washed with PBS and incubated overnight at 4°C with the appropriate primary antibodies, as shown in Table 1, at their appropriate dilutions. The protein expression against specific antibody was measured by incubating the samples with corresponding secondary antibodies coupled to Alexa Fluor-488 and Alexa Fluor-594 (Molecular Probes) for 1 h at room temperature. These fluorescent samples were counterstained with DAPI and mounted on glass slides using ProLong Gold antifade (Thermo Fisher Scientific, Waltham, MA, USA) mounting medium. Fluorescent images were analyzed under an Olympus X100 confocal microscope (Olympus).
Growing cattle-derived intestinal organoids were cultivated in a Matrigel dome as previously described [12]. To harvest the organoids, a 10× volume of enzyme-free cell disassociation buffer (1 mL) was added to a Matrigel dome (100 μL) in each well and incubated for 10 min in an incubator. To generate apical-out intestinal organoids, the organoids were then collected by centrifugation at 200×g for 5 min. The organoids were re-seeded and cultivated in ultralow-attachment 24-well plates with intestinal human organoid medium (Stem Cell Technologies) through suspension culture without Matrigel matrix. The morphology of the apical-out intestinal organoids was monitored daily under a microscope to evaluate polarity reversal [8].
FITC-dextran (4 kDa) (Sigma-Aldrich) was diluted in nuclease-free water and resulted in a 1 mg/mL working solution to evaluate epithelial barrier function. Growing cattle-derived basal-out and apical-out intestinal organoids were placed in 24-well plates. Then, 25 ng/mL FITC-dextran was added to each well, and the plate was incubated under normal growth conditions. The permeability was monitored using luminal absorption and recorded at 30-min intervals for up to 3 h under a Leica CTR6000 fluorescence microscope (Leica, Wentzler, Germany). The fluorescence intensity was calculated using ImageJ software.
Total RNA was isolated from prepared samples, including intestinal organoids, using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) as described previously [13,14]. The RNA quality and quantification was assessed by an Agilent 2100 bioanalyzer using an RNA 6000 Nano Chip (Agilent Technologies, Amstelveen, The Netherlands).
Quantitative RT-PCR was performed to assess the expression of several markers regarding intestinal stem cells and epithelium in ileum and jejunum tissues and growing cattle-derived intestinal organoids. Total RNA (1 µg) was reverse transcribed using the Superscript III First-Strand Synthesis System (Invitrogen, Waltham, MA, USA). The PCR mixture was prepared by adding 2 μL 10 pmol of each forward and reverse primer, 7 μL nuclease-free water, 10 μL SYBR Green qPCR Mater Mix, and 1 μL cDNA to a final volume of 20 μL. PCR was performed as follows: initial incubation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Sequence-specific products were identified by generating a melting curve. The Ct value represents the cycle number at which a fluorescent signal increased to a level significantly higher than the background, and relative gene expression was determined by the 2−ΔΔCt method [15]. The qPCR primers for each target gene and 18S ribosomal RNA (rRNA) that were used as previously described (Table 2). Gene expression levels were normalized to that of bovine 18S rRNA and performed using the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Waltham, MA, USA).
All data are expressed as the mean ± standard error from three independent experiments. The significance between groups was analyzed by two-way ANOVA of variance or Student’s t-test using GraphPad Prism V 6.0 software (GraphPad, San Diego, CA, USA). The differences were considered statistically significant at p < 0.05.
RESULTS AND DISCUSSION
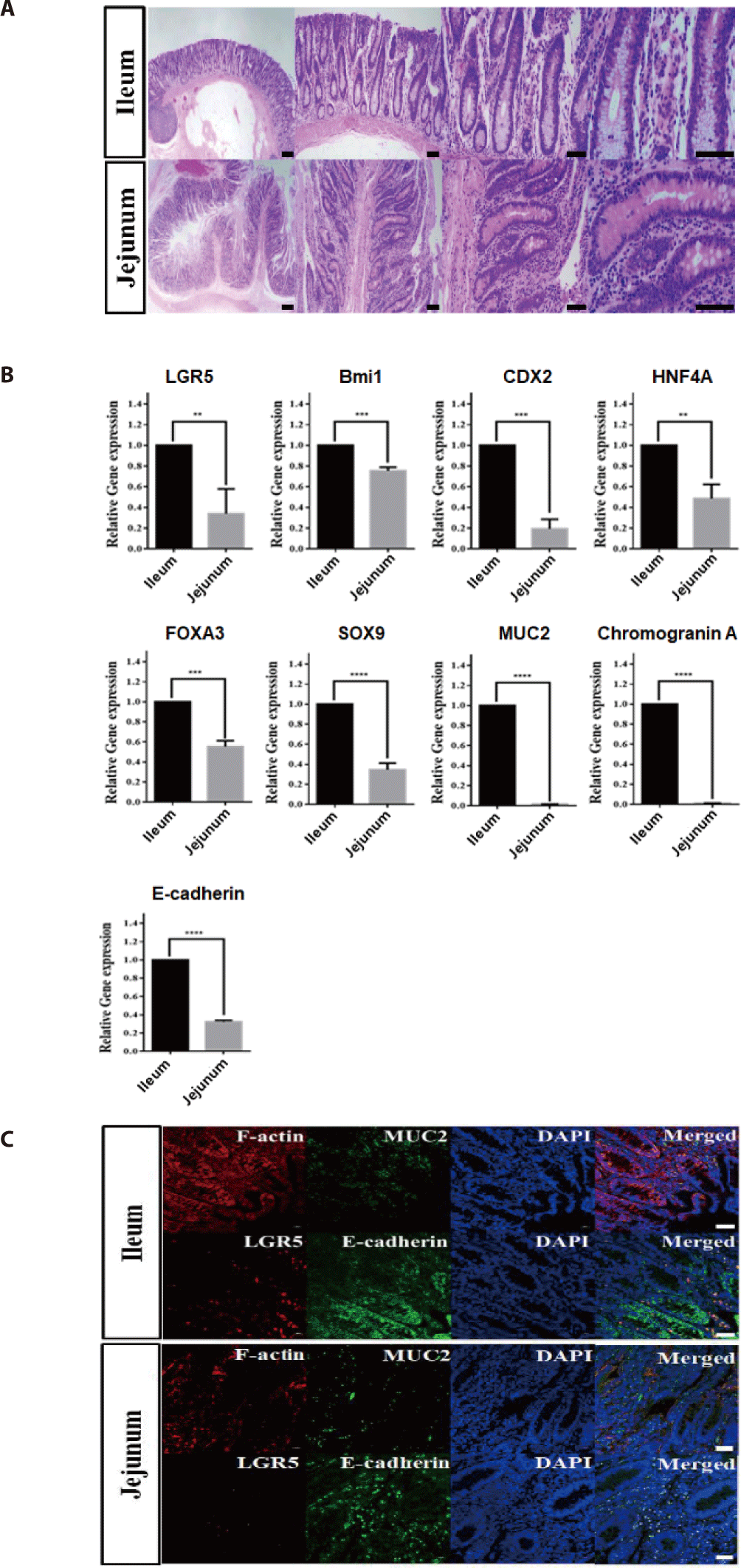
First, we selected ileum and jejunum fragments from the small intestine to identify suitable sites from which to more effectively isolate intestinal crypts of growing cattle (12 months old). Intestinal ileum and jejunum tissues were histologically analyzed using hematoxylin and eosin staining to identify distinct crypt and villus structures, including intestinal stem cells. Detailed views of vertical and horizontal sections from ileum and jejunum tissues showed the integral structures of the intestinal epithelium gland, such as crypts at the bottom and finger-shaped villi on the apical side, indicating the possibility of growth potentials for derivation of intestinal organoids from both the jejunum and ileum in growing cattle (Fig. 1A). In addition, to investigate the genetic properties of ileum and jejunum tissues, quantitative RT-PCR was performed. As shown in Fig 1B, intestinal stem cell-related genes, such as LGR5 (p < 0.01), Bmi1 (p < 0.001), CDX2 (p < 0.001) [16], HNF4A (p < 0.01) [17], FOXA3 (p < 0.001) [2], and SOX9 (p <0.001) [18], and intestinal epithelium-related genes, such as MUC2 (p < 0.001), Chromogranin A (p < 0.001), and E-cadherin (p < 0.001), were expressed at significantly higher levels in the ileum than in the jejunum. Furthermore, to identify intestinal stem cells of the ileum in vitro, immunohistochemical analysis of several markers related to intestinal stem cells and epithelial cells was conducted. As shown in Fig. 1C, the ileum of the small intestine exhibited distinct expression pattern, which included leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), which was found to be expressed with F-actin in the intestinal epithelial cytoskeleton. Moreover, the fluorescently stained villi and crypt structures showed epithelium-specific expression of MUC2 in goblet cells and E-cadherin in adherent junctions, indicating that the authenticity of intestinal stem cells and epithelium in the ileum of the small intestine in growth cattle. However, the jejunum had a relatively low expression compared to the ileum of the small intestine. Collectively, these results strongly suggested that the ileum of the small intestine had the most growth potentials for the derivation of intestinal organoids from growth cattle.
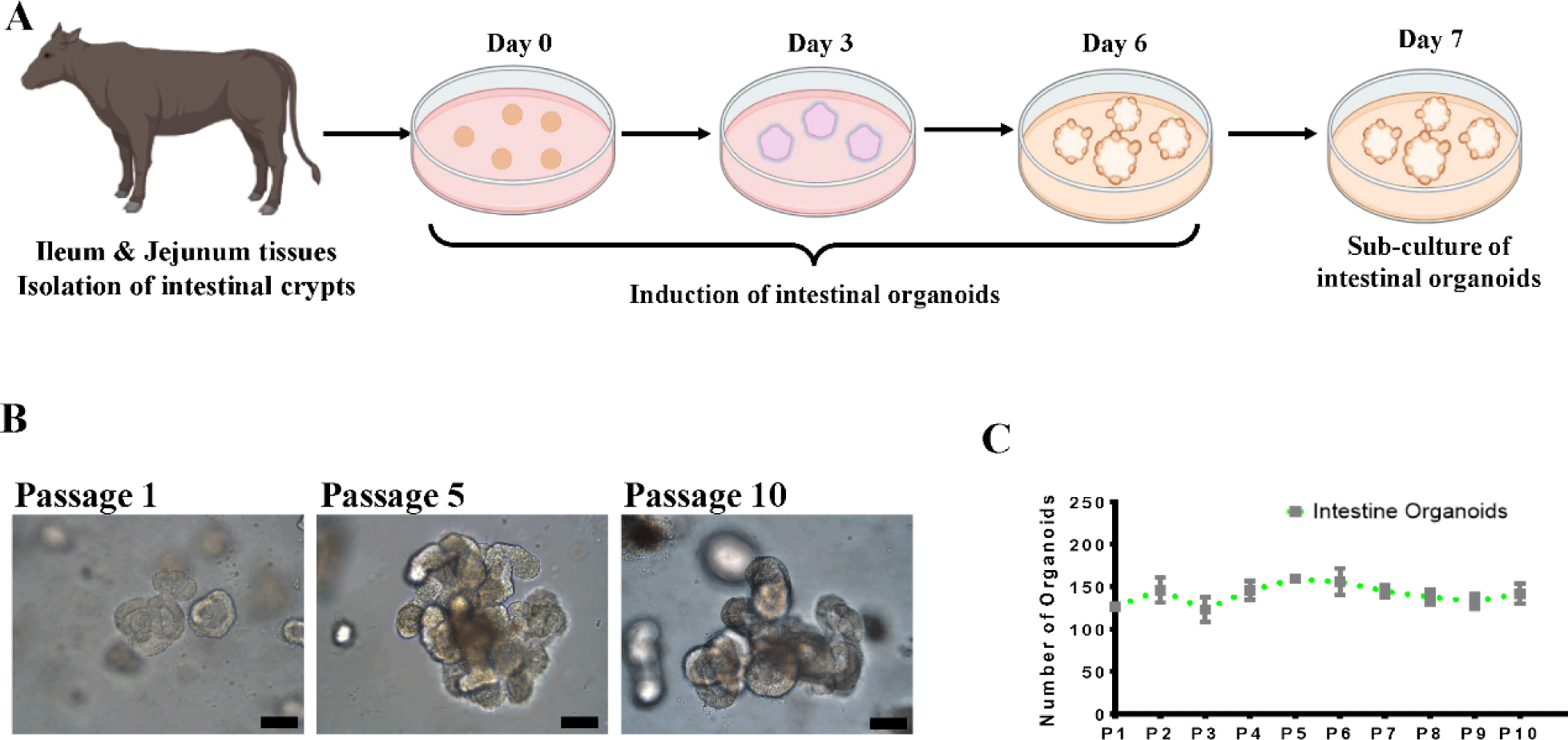
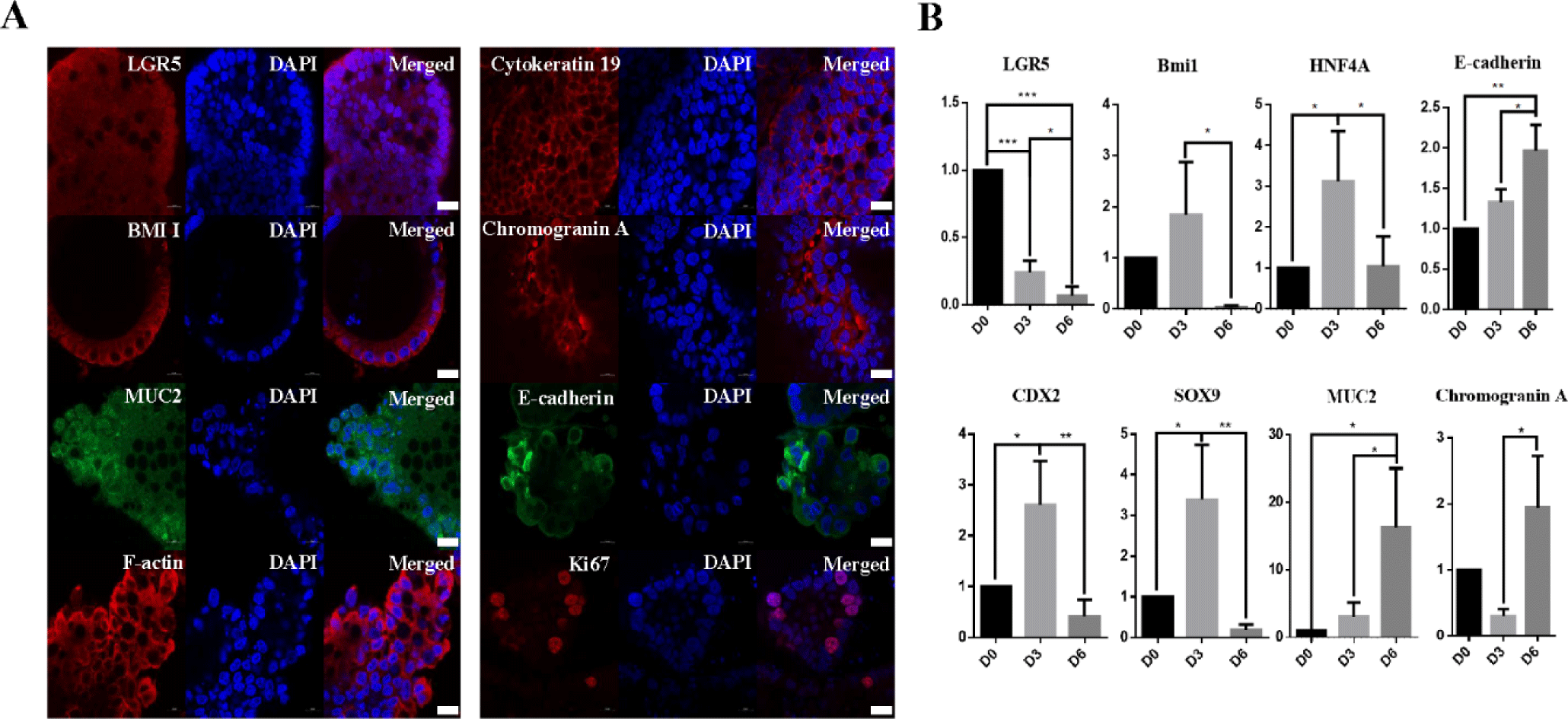
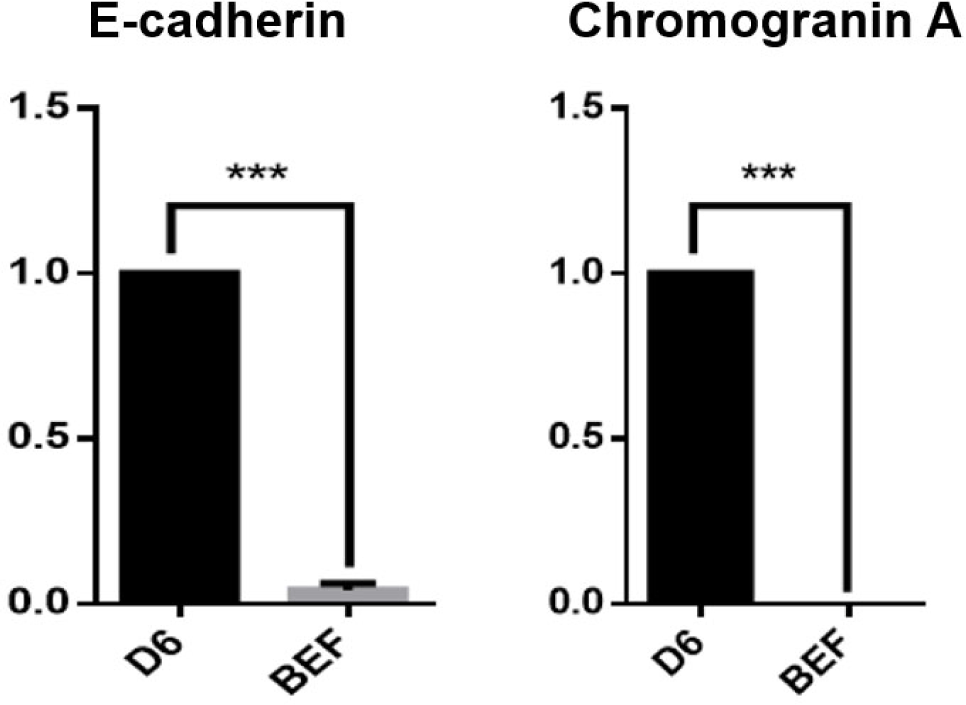
Intestinal crypts were isolated from the small intestine (ileum and jejunum) of healthy growth cattle (12 months old), and cultivated in IntestiCult medium at Matrigel dome. Fig. 2A illustrates the experimental procedure for the isolation of growth cattle-derived intestinal crypts, including the induction and cultivation of intestinal organoids. In this study, we attempted to produce intestinal organoids by isolating crypts from the jejunum and ileum of the small intestine to generate organoids specific to different regions of the small intestine. Interestingly, although there was no particular difference between the jejunum and ileum in growing cattle in terms of crypt and villus structure, intestinal organoids were confirmed to be formed by ileum (Fig. 2B). However, organoid formation did not occur jejunum. According to our previous results, intestinal organoids were described to exhibit detailed structures, such as spheroidal, stomatocyte, budded/elongated and branched structures, at each passage from Day 0 to the fully mature structure observed on Day 7 [2,19]. As shown in Fig. 2B, these organoids showed various morphologies, such as spheroidal (round shaped) and mature villi and crypt-like structures, from the P1 to P10 generations at early passages. Furthermore, they showed stable growth for more than 10 passages with an average of 130–150 organoids present in the Matrigel dome at each generation, indicating the continuous proliferation and recapitulating capacity of the ileum-derived intestinal organoids (Fig. 2C). These results were consistent with establishment of the jejunum-derived intestinal organoids from adult bovines [2]. To more characterize the cellular potential of the ileum-derived intestinal organoids (P5) in growing cattle, we investigated the spatial expression of several specific markers of in intestinal stem cells and the epithelium. Intestinal organoids do not consist of only one cell but consist of various cells, such as stem cells, Paneth cells, and intestinal endocrine cells [10,20]. Consistent with previous results [2], the ileum-derived intestinal organoids had distinct patterns of gene expression, such as LGR5 and Bmi1 expression. Moreover, the fluorescently stained organoids showed epithelium-specific expression of Mucin2 in goblet cells, which contributes to epithelial barrier integrity, E-cadherin in adherent junctions, F-actin in the intestinal epithelial cytoskeleton, Chromogranin A in enteroendocrine cells, and Cytokeratin 19 in enterocytes, suggesting that the organoids closely mimicked the in vitro organ physiology (Fig. 3A). In addition, we investigated the genetic expression patterns of the ileum-derived intestinal organoids through expression profiling. As shown in Fig. 3B, the expression of intestinal stem cell-related genes, such as LGR5 (p < 0.001), was significantly higher in the ileum-derived intestinal organoids on Day 0 than on Day 3 and Day 6, while the expression of intestinal stem cell-related genes, such as Bmi1 (p < 0.05), CDX2 (p < 0.05), HNF4A (p < 0.05) and SOX9 (p <0.05), was significantly higher in the ileum-derived intestinal organoids on Day 3 than on Day 0 and Day 6. In addition, the expression of intestinal epithelial markers, such as MUC2 (p < 0.001), Chromogranin A (p < 0.05) and E-cadherin (p < 0.01) was significantly higher in the ileum-derived intestinal organoids on Day 6 than on Day 0 or Day 3. Moreover, the expression of intestinal epithelial markers, such as Chromogranin A (p < 0.001) and E-cadherin (p < 0.001) was specifically expressed in the ileum-derived intestinal organoids on Day 6 than bovine embryonic fibroblast (BEF) (Fig. 4). This confirmed the development and differentiation of intestinal stem cells into intestinal organoids over time. Taken together, these results demonstrated that the genetic expression patterns and cellular potentials of the ileum-derived intestinal organoids were highly similar to those observed in vitro.
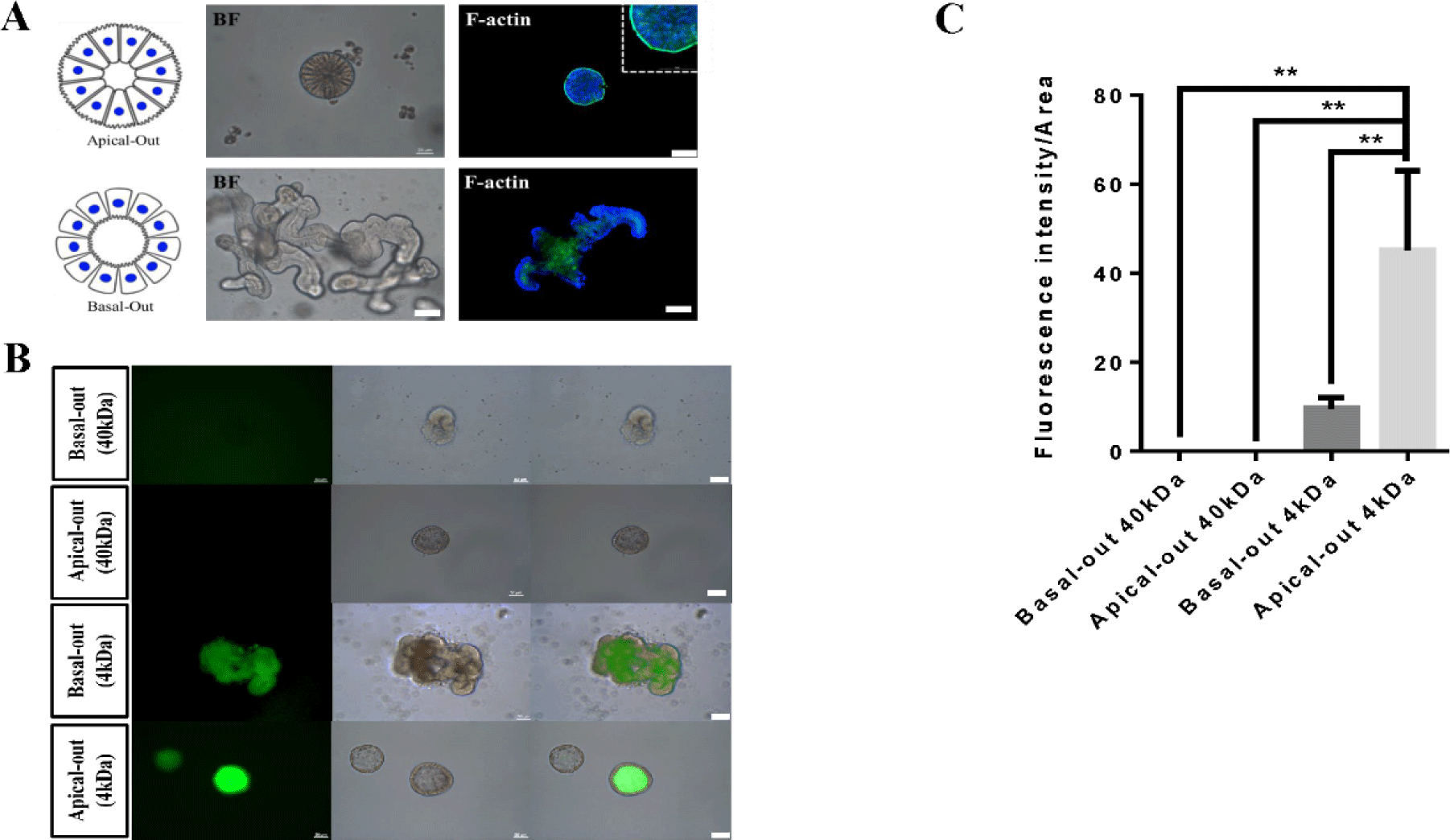
Recently, intestinal organoids have been considered potential alternatives to in vitro systems and have become the focus of research about livestock, including cattle, pig and chicken [1,11,21,22]. In particular, intestinal organoids have been described as the ideal model of mucosal permeability since they that can model the absorption of bacteria, viruses, and nutrients [7,9,23–26]. However, there are practical limitations associated with the potential use of intestinal organoids, such as host-pathogen interactions and nutrient absorption, because intestinal organoids cultivated in Matrigel have all basal-out structures [8–10]. To overcome the limitations of current growing cattle-derived basal-out intestinal organoid models cultivated in Matrigel matrices, we generated apical-out intestinal organoids through suspension culture without Matrigel matrix, showing that these organoids exhibited an apical-out membrane on their surfaces (Fig. 5A). From our results, on day 3 of culture in suspension without the Matrigel matrix, the apical-out form began to emerge. In addition, we investigated the spatial expression of a specific marker, F-actin, associated with epithelial characteristics in apical-out-derived intestinal organoids derived from growing cattle. Based on our results, it was confirmed that F-actin was strongly expressed on the surface, showing that intestinal epithelial cells were inverted and formed differently compared with those observed in basal-out organoids (Fig. 5A). The intestinal epithelium plays an important role in the absorption of nutrients through the membrane and diffusion of small molecules across the intestinal barrier [27,28]. Furthermore, we tested the paracellular permeability of the epithelial layer using fluorescent tracers. We previously reported that FITC-dextran 4 kDa did not reach the apical surface of bovine intestinal organoids due to their basal-out structures [2]. Generally, intestinal pathogens penetrate the apical membrane of intestinal epithelial cells [8]. However, apical-out bovine intestinal organoids overcame this limitation. Markedly, they showed the key functionality with regard to a high permeability for compounds up to FITC-dextran 4 kDa in size, while FITC-dextran 40 kDa failed to enter the organoid lumen because of intestinal epithelial barrier integrity. In addition, when the fluorescence of the apical-out and the basal-out structure were compared, it was confirmed that the transmittance of the apical-out structure was significantly higher and were better than other models (Figs. 5B and 5C), indicating apical-out intestinal organoids is the best model to study for various purposes. Together, these functional testing results suggested that apical-out intestinal organoids were physiologically relevant and exhibited properties similar to those of the gut in vitro. Finally, these novel organoids may be useful for evaluating nutrient absorption capacity and in experiments on intestinal epithelial cells.
CONCLUSION
In this study, we successfully established the ileum-derived intestinal organoids from growing cattle and generated a more reliable research model for intestinal organoids by producing organoids with apical-out structures and characterized their cellular potentials. Growing cattle-derived intestinal organoids have potential for use in various purposes in the field of animal biotechnology, such as disease modeling and feed efficiency measurement.
















