INTRODUCTION
Global meat production is being increased, estimated to 337.2 million tons in 2020 [1]. Furthermore, following the COVID-19 pandemic, meat production is expected to rise to 373 million tons by 2030 [2]. According to the global trend in 2018, 127.31 million tons of chicken, 4.46 million tons of duck, 120.88 million tons of pork, and 71.61 million tons of beef were consumed among meat groups [3]. In addition, beef and buffalo meat accounted for about 22% of meat production from 1961 to 2018, which was reduced by half, but pork production remained constant at approximately 35%–40% [3]. Landrace is a large white pig crossbred from Denmark. This breed requires a slower feeding cycle but has a faster growth rate [4]. The Yorkshire breed, which originates from England, has good muscles not only in the pork belly, but also in the thigh due to its good growth rate and long body. Hence, many countries have used this type as a crossbreed to produce meat [5]. The Duroc breed originated from the United States shows a greater capacity of accumulating intramuscular fat (IMF), which shows better eating quality, flavor and consumer preference [6,7]. The crossbreed of Landrace, Yorkshire, and Duroc (LYD) is raised as a meat-producing pig because of its excellent fertility and high-quality meat [8]. The Korean traditional pig, a black and short-haired breed, was bred since the 1970s [9,10]. Compared to the LYD crossbreed, it has a lower performance, but a stronger disease prevention capacity [10], but has a better texture and harder fat than the LYD crossbreed [11]. Woori black pig (WB) was developed to improve meat quality and growth efficiency by crossbreeding Duroc with Korean native black pig [12]. Among factors that affect the meat quality including juiciness, color, pH, IMF, and sarcomere length, sarcomere length is positively related to the meat quality or marbling score [13]. Another important factor muscle fiber type, which is categorized into several different types depending on myosin heavy chains (type I, II, IIa, IIx, or IIb), energy metabolism (oxidative or glycolytic), or speed of contraction (fast or slow), affects muscle growth and development as well as meat quality [14]. Skeletal muscle satellite cells (SMSC), also known as muscle stem cells located between the basal lamina of muscle fibers and the fascia, play crucial roles in skeletal muscle hypertrophy and regeneration [15]. Characterization of SMSC, therefore, can indirectly provide the capacity of muscle growth, development, and metabolism ultimately affecting the animal growth efficiency and meat quality [16,17]. There are several myogenic genes including Myosin heavy chain (MyHC), paired box gene 3 (Pax3), myoblast determination protein 1 (MyoD), and myogenin [18]. MyHC defines the fiber type and contraction speed [19]. Myostatin, encoded by the MSTN gene inhibits the growth and differentiation of muscle cells [20]. Pleomorphic adenoma gene 1 (PLAG1) induces cellular growth and in some cases it is related to specific cancers [21]. Adipogenic genes include fatty acid synthase (FASN) a key enzyme related to the synthesis of fatty acids. Stearoyl-CoA desaturase-1 (SCD), which inhibits the growth of adipocytes, and Peroxisome proliferator activated receptor-γ (PPAR-γ), the major regulator involved in differentiation of adipocytes [22]. CCAAT/enhancer-binding protein (CEBP) exerts its functions during the initial stage of adipogenesis, and adiponectin plays a role in body fat reduction [23]. Although growth rate and meat quality of WB has been investigated [24], there has not been many studies which compare and analyze the growth and differentiation of muscle cells and relevant gene expression. The current study therefore compares the number of SMSC and related gene expression patterns of muscle and fat tissues of LYD to those of WB.
MATERIALS AND METHODS
Dulbecco’s Modified Eagle Medium-F12 (DMEM-F12), fetal bovine serum (FBS), horse serum (HS), and antibiotic-antimycotic (AA) were purchased from Gibco (Thermo Fisher Scientific, Waltham, MA, USA). Trizol reagent (AccuzolTM Total RNA Extraction Reagent, Bioneer, Seoul, Korea), diethylpyrocarbonate (DEPC) water (Bioneer), cDNA transcription kit (AccuPower CycleScript RT PreMix, Bioneer), qPCR MasterMix (Bioneer), and nuclease-free-water (Ambion®, Austin, TX, USA) were purchased for RNA extraction, cDNA synthesis, and quantitative real-time polymerase chain reaction (PCR). The experimental protocols for this research were reviewed and approved by the Institutional Animal Care and Use Committee at the National Institute of Animal Science (NIAS-2020-437).
The WB and LYD sire pigs were provided by the National Institute of Animal Science, Rural Development Administration (RDA), Korea. Animal sampling methods were followed upon ethical clearance. The pigs were slaughtered for muscle sample collection at RDA.
A total of 24 pigs (12 LYD and 12 WB) were used in this study. Three pigs in each breed were sacrificed when their body weights (b.w.) reached 50 ± 0.20, 75 ± 2.89, 100 ± 1.78, and 120 ± 2.37 kg of LYD, and 50 ± 1.31, 75 ± 1.30, 100 ± 2.20, and 120 ± 0.95 kg of WB. Muscle and adipose tissues were obtained from the hind leg. Skin of hind leg was sterilized with 70% ethanol, subcutaneous fat sampled. Tendons were transected at both proximal and distal sides while holding one tendon with forceps and transecting the other tendon with the scalpel and femoris muscle was harvested.
SMSC were isolated from the femoris muscle using Pronase enzyme digestion followed by centrifugation at 1,200×g and cells were then seeded at 2×104 cells/mL in a T-25 flask, cultured in growth media consisting of DMEM-F12 containing 1% of AA and 10% of FBS. After 24 hours, the media was replaced with fresh growth media, and the number of cells was counted on day 1. Then, the media was replaced with fresh media every 48 hours. Trypsinization for cell counting was performed using 0.05% of Trypsin-EDTA (Gibco, Gaithersburg, TN, USA) on days 1, 3, 5, and 7. The number of cells were counted using a hemocytometer (Counting Chamber, Paul Marienfeld GmbH & Co., Am Wöllerspfad, Germany). This was performed in triplicate for each sample and averaged each day. Population doubling time (PDT) was calculated by PDT Calculating software.
The total RNA was isolated using Trizol reagent (ACcuzolTM Total RNA Extraction Reagent, Bioneer). Specifically, the Trizol reagent was used at a concentration of 1 mL per 1 g of muscle or adipose tissue sample. Next, 200 µL of chloroform per 1 mL of Trizol reagent was added and the sample was shaken vigorously for 15 seconds. The mixture was then incubated on an ice block rack for 5 minutes and centrifuged at 12,000×g for 15 minutes at 4°C. Then, take off the upper aqueous phase and transferred to a new tube where an equal volume of isopropyl alcohol was added. The sample was then mixed and incubated at −20°C for 10 minutes. Next, the sample was centrifuged at 12,000×g for 10 minutes at 4°C. The supernatant was then removed upon which 1 mL of 80% ethanol was added. The sample was mixed well and then centrifuged at 12,000×g for 5 minutes at 4°C. The supernatant was then removed, and the pellet was dried. Finally, the pellet was resuspended in 20 µL of DEPC water (Bioneer) and the purity and RNA concentration were checked for use in cDNA synthesis using a Microplate Spectrophotometer (Multiskan Sky, Thermo Fisher Scientific) and µDrop plate (µDropTM, Thermo Fisher Scientific). The concentration of the obtained RNA samples was then adjusted to 1 µg. The RNA samples were then reverse transcribed into cDNA using the cDNA reverse transcription kit (AccuPower CycleScript RT PreMix, Bioneer) with a GeneAmp PCR System 9700 machine (Applied Biosystem, Singapore). RNA samples were added to the PreMix tub, which was filled with 20 µL of DEPC water. The machine was then operated based on the cDNA synthesis condition, which was composed of a synthesis step at 45°C for 60 minutes and a heat inactivation step at 95°C for 5 minutes.
The cDNA was used in the comparative cycle threshold experiment for relative gene expression. A quantitative real-time PCR was performed using the AccuPower® 2X Greenstar qPCR MasterMix (Bioneer) and StepOnePlus Real-Time PCR system (Applied Biosystems). First, the cDNA concentration was adjusted to 100 ng/µL with nuclease-free-water (Ambion®). Then, the 20 µL total volume was made by mixing 1 µL of diluted cDNA, 10 µL of the master mix (containing fluorescent material), 0.5 µL of the forward primer, 0.5 µL of the reverse primer, and 8 µL of nuclease-free-water. The mixtures were then stored in reaction tubes (MicroAmp® Fast Reaction Tubes, Applied Biosystems). qPCR reaction conditions were implemented based on the following protocol. During the first stage, the initial denaturation was completed at 95°C for 5 minutes. Denaturation was then performed at 95°C for 15 seconds. The next step involved 40 repeated cycles (the annealing temperatures and times are shown in Table 1), and an extension at 72°C for 45 seconds. The last step was the melt curve, which was included in the final extension and hold phase. The final extension was performed at 95°C for 15 seconds, 60°C for 1 minute, and 95°Cfor 15 seconds. During the last hold stage, the temperature remained at 4°C for infinity. The primer sequences of the housekeeping and target genes are listed in Table 1.
Each sample was 1 × 1 × 1 cm cut in the orientation of the muscle fiber on a flat surface and stored at -85°C in a deep freezer (TSE320GPD, Thermo Fisher Scientific). The frozen sample was cut into 10 μm sections at -25°C with a cryostat cryocut micro-tome (CM3050 S, LEICA®, Germany), then the sarcomere length was observed under a high- resolution field emission scanning microscope (MIRA3-LM, Tesan, Czech Republic). The muscle fiber cross section area was measured using a laser confocal scanning microscope (ZEISS LSM 800, ZEISS, Oberkochen, Germany).
Statistical analysis was performed using Prism 9.4.0 (GraphPad). The data are presented as mean ± standard deviation. Two-way analyses of variance (ANOVA) followed by Tukey significant difference test were performed. Significant differences were considered by p< 0.05 (*p< 0.05, **p< 0.01, and ***p< 0.001) in all Figures.
RESULTS AND DISCUSSION
Result of growth performance of the LYD and WB is shown in Table 2. The initial and final b.w. of the LYD crossbreed were 26.20 ± 0.60 kg and 124.37 ± 2.37 kg, those of WB were 24.86 ± 2.40 kg and 120.63 ± 0.95 kg, respectively, with no significant difference. Average daily gain, however, was greater in LYD crossbreed than WB (975.70 ± 105.15 g vs. 768.49 ± 69.17 g; p< 0.001). Daily feed intake was not significantly different between 2 groups (2,736.04 ± 120.35 g vs. 2,736.52 ± 131.72 g; p=0.974).
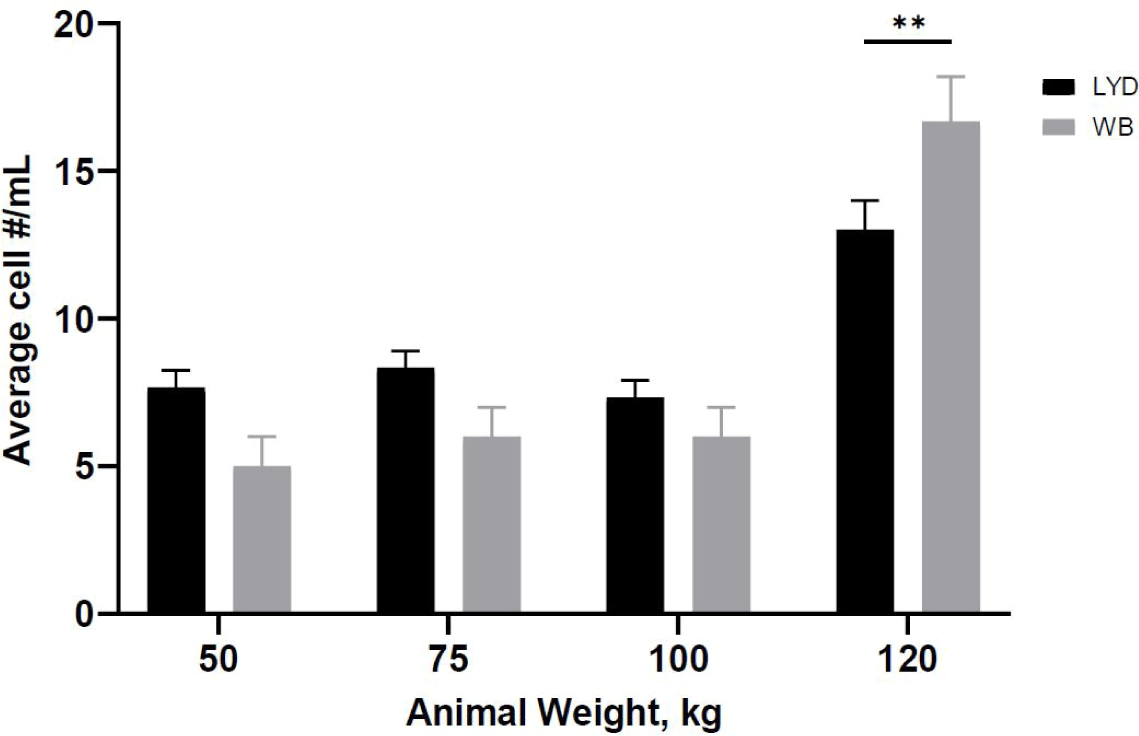
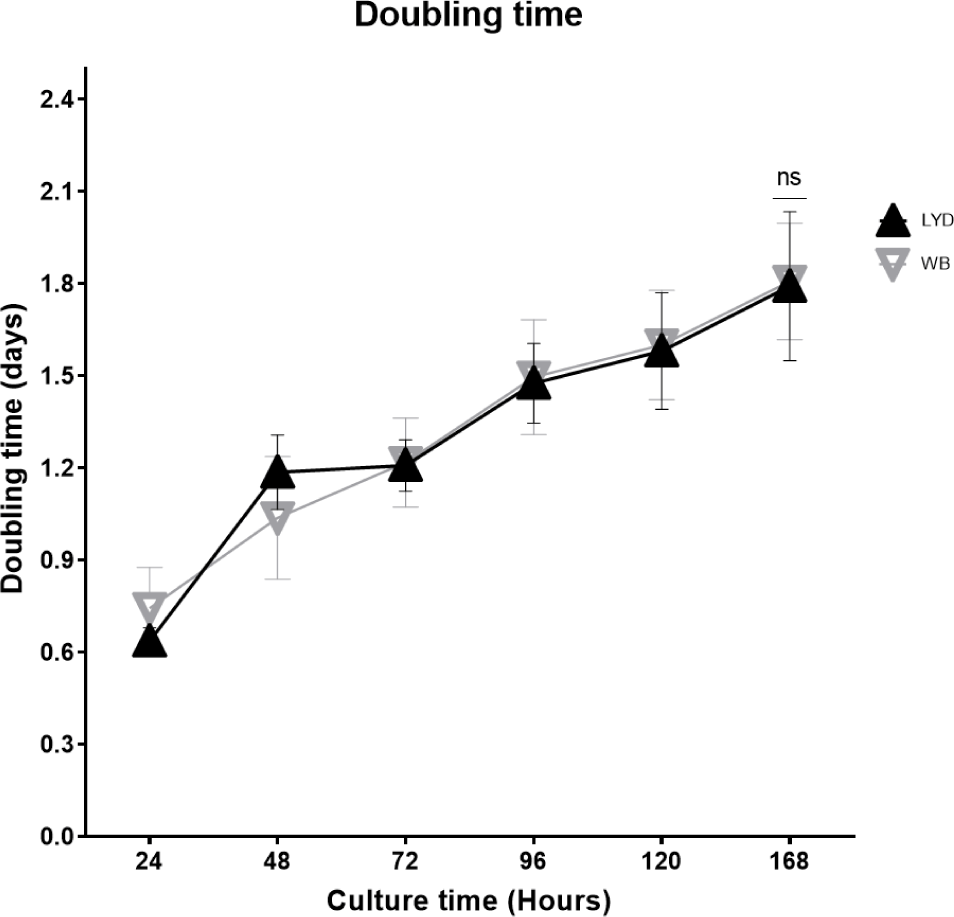
Number of cells harvested, growth rate and related gene expression is crucial to understand the mechanisms by which genetic traits regulate the muscle growth and development [16]. Average muscle cell yields were tended to be higher in LYD than WB at 50, 75, or 100 kg b.w. groups, but WB at 120 kg showed more cells than LYD (p < 0.01) (Fig. 1). Although there was a difference in SMSC number between LYD and WB, cell morphology and doubling time was not different (Figs. 2 and 3). Cell doubling time refers to the time taken to double the number of cells, and the difference can stem from various reasons including breeds, age, genetic traits, gender of animal, or cell culture condition. To this end, genes related to myogenesis were analyzed.
Cell number is increased through the proliferation process. With external stimuli and growth factors, muscle forms its structure from multiple elongated muscle fibers via cell fusion or differentiation [25]. This developmental process involves many factors and genes [26]. During maturation, orchestration of these gene expressions render the myoblasts fused into myotubes and matured [27]. MyoD acts as a transcriptional activator and thus is engaged in muscle hyperplasia and hypertrophy. Myogenic factor 5 (Myf5) and myogenin (MyoG) are involved in promoting gene expression and muscle development [28]. MyoG accelerates the transcription of muscle-specific genes. The gene expression pattern in skeletal muscle tissue shows that MyoD is expressed throughout all stages [29]. As shown in Fig. 4A, muscle from LYD crossbreed had a greater expression levels of MyoD, MyHC, and Pax3 than the WB. In particular, MyoD expression was higher in LYD than in WB at 50, 75, and 100 kg b.w. groups (p < 0.001).
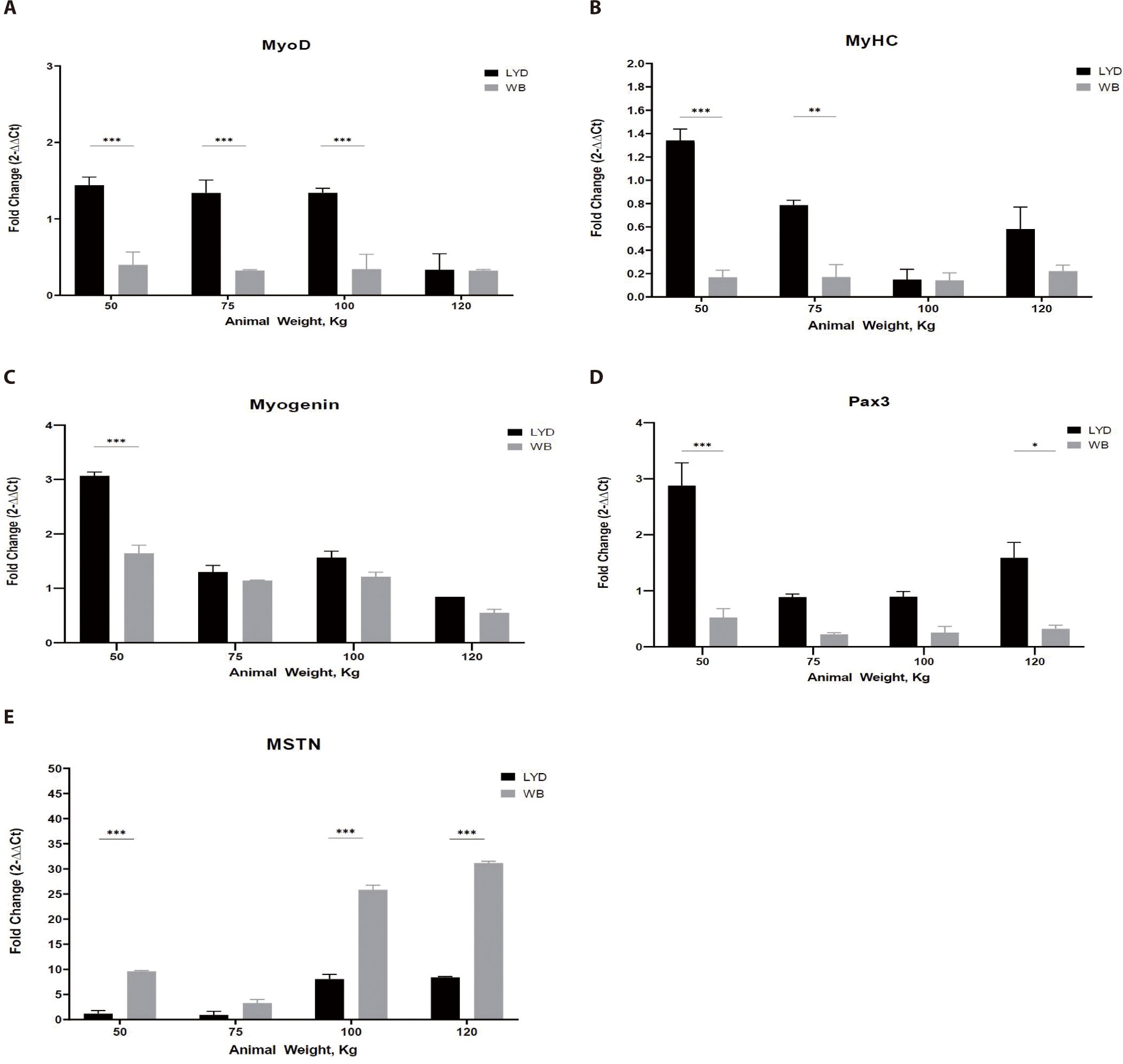
MyHC is a major structural protein in muscle that converts chemical energy into mechanical energy through the hydrolysis of ATP and expressed in proportion to the amount of muscle [30]. In Fig. 4B, muscle from LYD showed a higher MyHC expression.
Pax3, a paired box transcription factor that regulates proliferation, migration, and cellular apoptosis, plays a key role in controlling myoblast fusion and skeletal muscle fiber development and differentiation [31]. Pax3 expression was higher in femoris muscles from LYD at b.w. of 50, 75, 100, and 120 kg compare to those from WB (Fig. 4C; p < 0.001). Based on its role in muscle development, relatively lower b.w. of WB pigs might be related to this lower Pax3 expression in the current study.
Myogenin is engaged in the functions of MyoD, Myf5, and myogenic regulatory factor 4 (MRF4) as a MyoD family [32]. Myogenin expressed more in muscle tissue from 50 kg LYD (Fig. 4D) compare to other groups (p < 0.001).
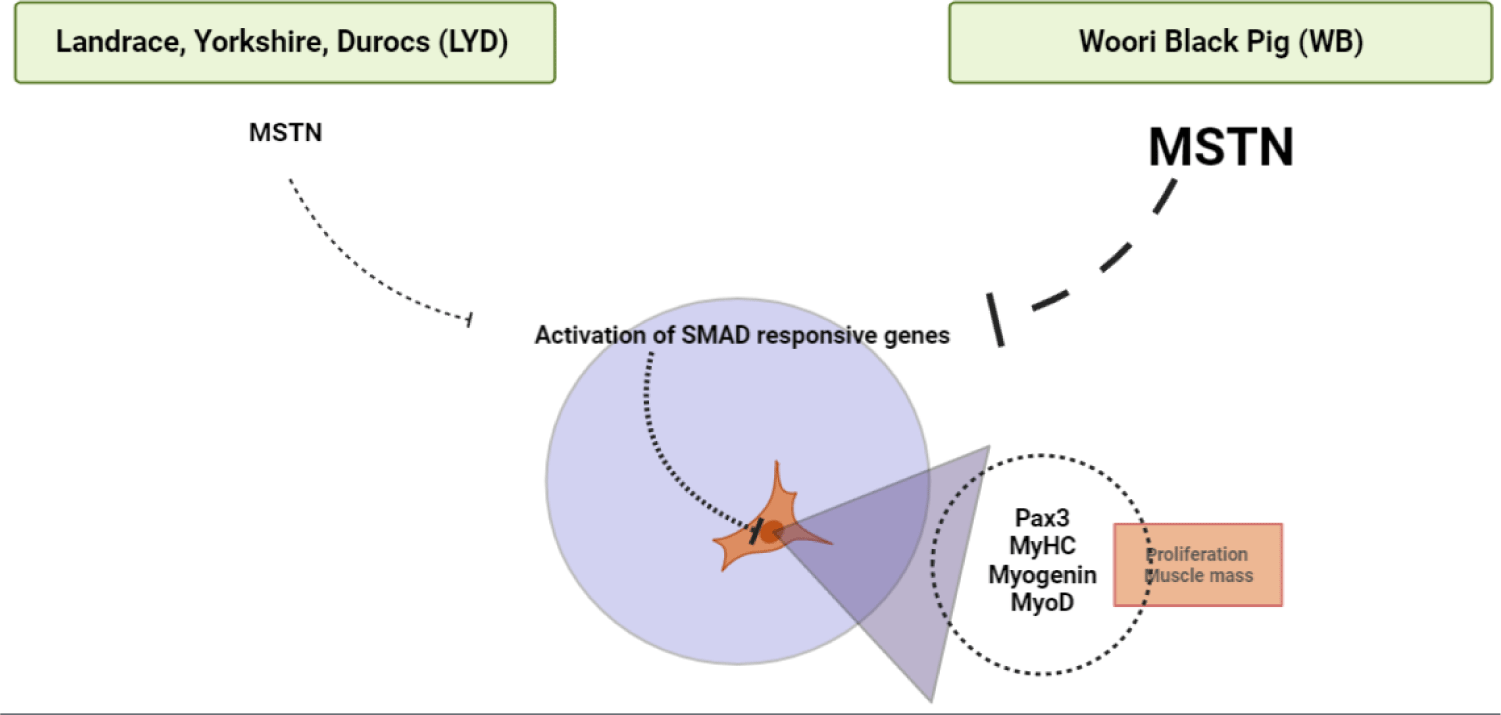
MSTN binds various transforming growth factor (TGF)-beta receptors that induce activation of SMAD sequence transcription factors down-regulating skeletal muscle cell proliferation and differentiation [33]. In Fig. 4E, MSTN expression was higher in WB group at 50, 100, 120 kg b.w. groups than in LYD (p < 0.001) which can partially explain the relatively lower daily gain in BW in the current study.
Adipogenesis is a process involving the development preadipocytes derived from multipotent mesenchymal stem cells (MSCs) [34]. It is a multistep process with the sequential activation of numerous transcription factors containing CEBP and PPAR-γ [35]. To reach maturity, these cells must go through three major well-defined steps: commitment of MSCs to the adipocyte lineage; somatic cell division and expansion involving DNA and cell duplication as well as terminal differentiation, including gene expression and transcription factors (such as CEBP and PPAR-γ); and an increase in lipid formation and the introduction of lipogenic genes, including acetyl CoA, carboxylase, FASN, and adipocyte fatty acid binding protein [36]. PLAG1 is an RNA-specific polymerase which regulates the activity of DNA-binding transcriptional factors and proximal promoter DNA-binding transcription, which induces the up-regulation of target genes, a higher proliferation rate, and transformation. Some stimulators include PPAR-γ, macrophage colony stimulating factor, prostaglandins, insulin-like growth factor I (IGF-l), glucocorticoids, and fatty acids [37]. CEBP-β is considered the most important factor and is induced rapidly after the induction of adipogenic stimuli [38].
As shown in Fig. 5A, LYD muscle from 75, 100, 120 kg showed greater CEBP-β expression (p < 0.001).
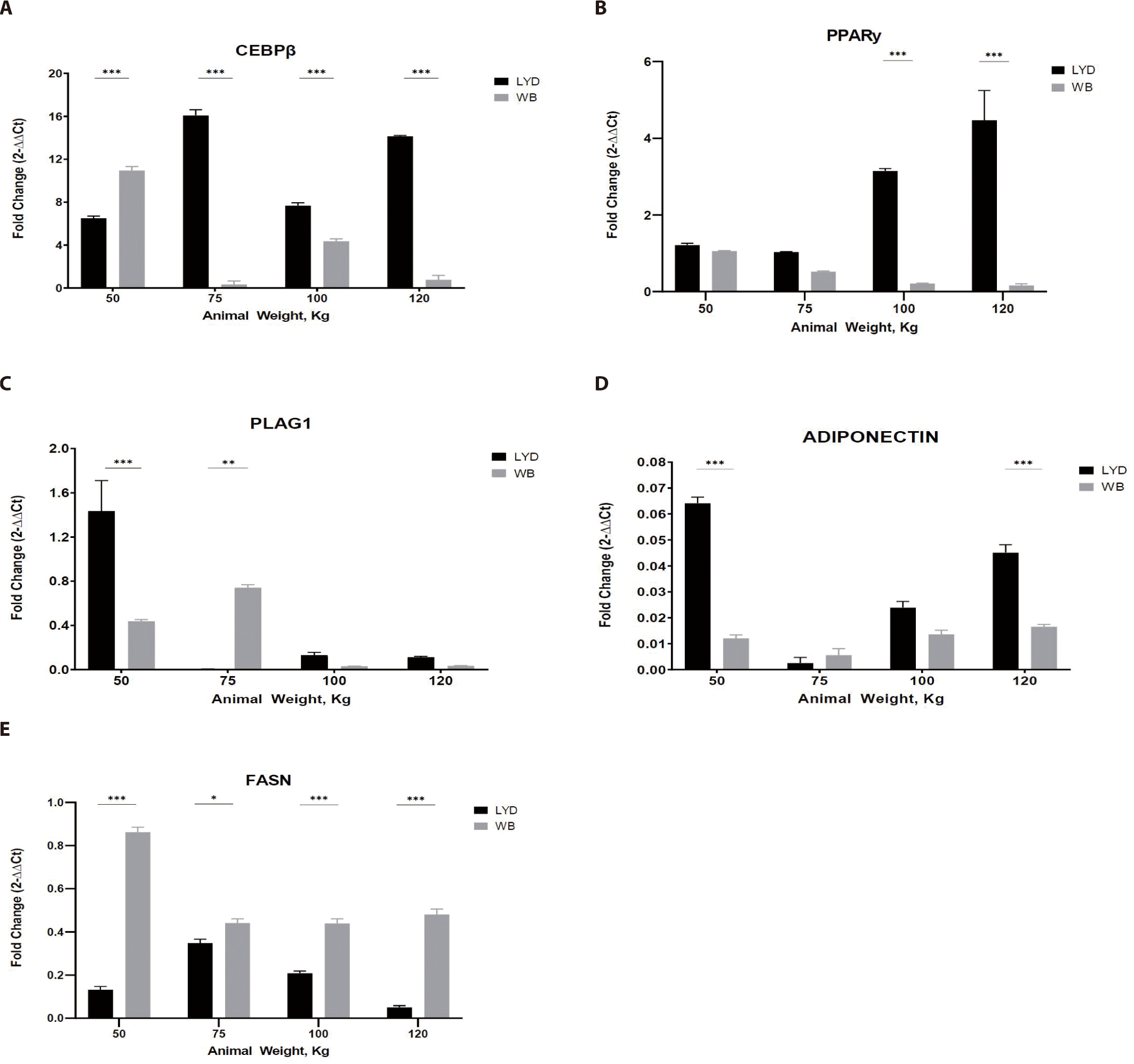
Adipose tissue from LYD at 75, 100, and 120 kg b.w. showed a higher expression than the WB (Fig. 5A; p<0.001). As a master regulator of fat cell differentiation, PPAR-γ is working with CEBP [39]. Gene expression pattern of PPAR-γ and CEBP was similar in Figs. 5A and 5B, but these patterns are not always mirroring the actual adipogenesis. Since their expression is precisely regulated in a time-dependent manner, expression levels of these genes can be up- or down-regulated very rapidly.
PLAG1 (Fig. 5C), known as the zinc finger transcription factor, has an association with muscle growth showing that growth of PLAG1 knock-out animals decreased by 50% compared to the normal group [40]. Expression of PLAG1 in the current study, however, is somewhat inconsistent and tended to decrease as b.w. increased in both breeds. Since gene expression is a rapid and complex process to cope with the environmental cues, integration of gene expression will be necessary to precisely understand this expression plasticity.
Adiponectin expression was greatest in muscle from 50 kg LYD among groups (Fig. 5D; p<0.001). Adiponectin plays a role in improving insulin resistance linked to body fat reduction [41], so, its expression pattern might mirror the speed of muscle growth. Further investigation is needed to elucidate this relationship.
Based on the expression pattern of FASN in Fig. 5E, more fat accumulation could be expected in WB compare to LYD since it is involved in fat synthesis [42].
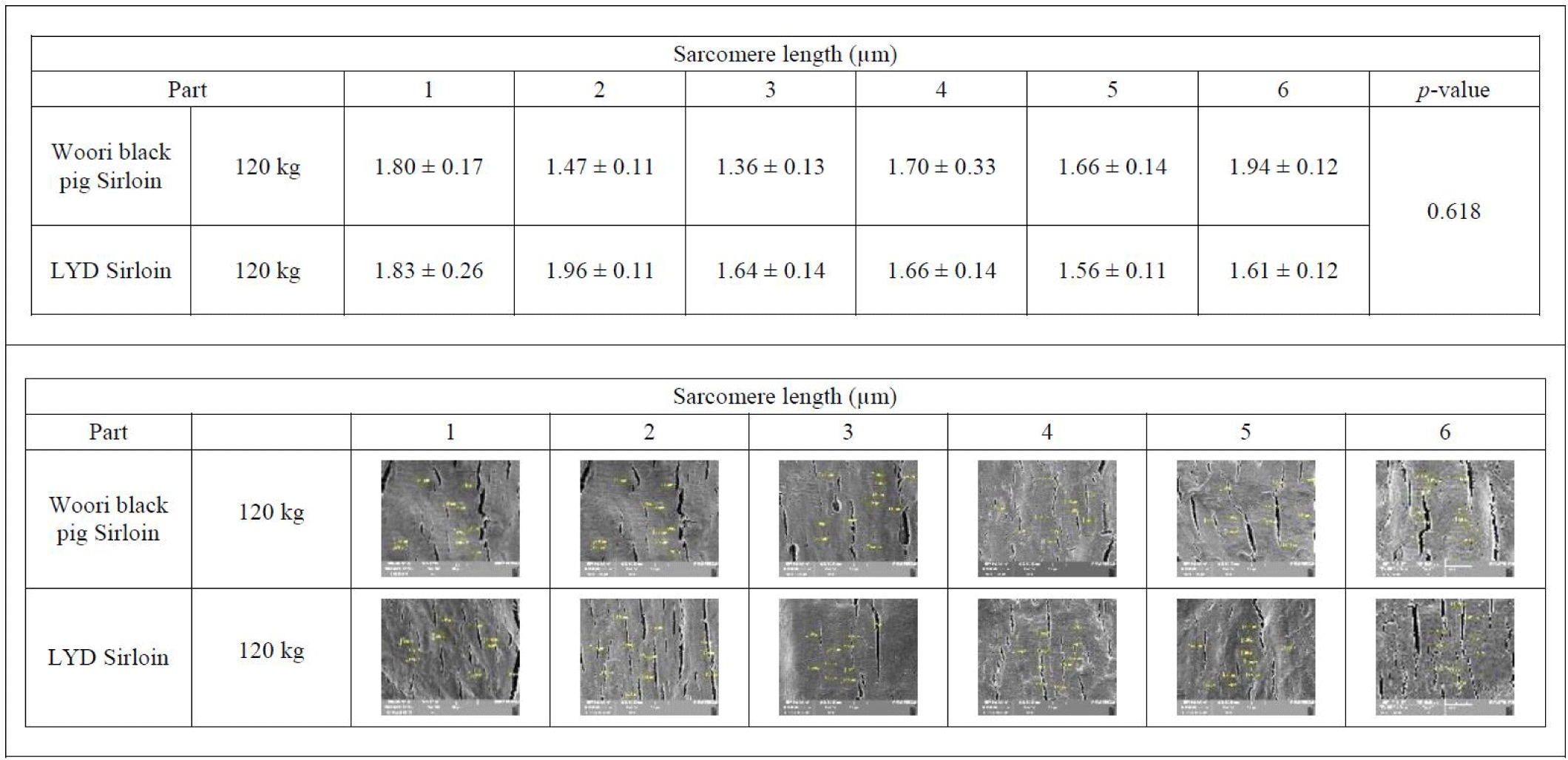
Sarcomere is a major player in muscular contraction and contains 28 or more proteins including myosin, actin, titin, tropomyosin, troponin, and nebulin. Among these, actin and myosin play an important role in muscle contraction together with tropomyosin and troponin. In the relaxed muscle of a living animal, the sarcomere length is estimated to be around 2.5 µm. Meanwhile, in the rigor mortis condition where the energy inside the muscles after death is exhausted, actin and myosin filaments cannot be detached from one another, and the corresponding sarcomere length is reduced by half the typical length [43]. Interestingly, a study conducted by Ward et al. indicated that sarcomere length is positively related to the marbling scores [44]. Average sarcomere length of muscles from WB and LYD were 1.710 μm and 1.655 μm, respectively with no statistical difference (p= 0.618) (Fig. 6).
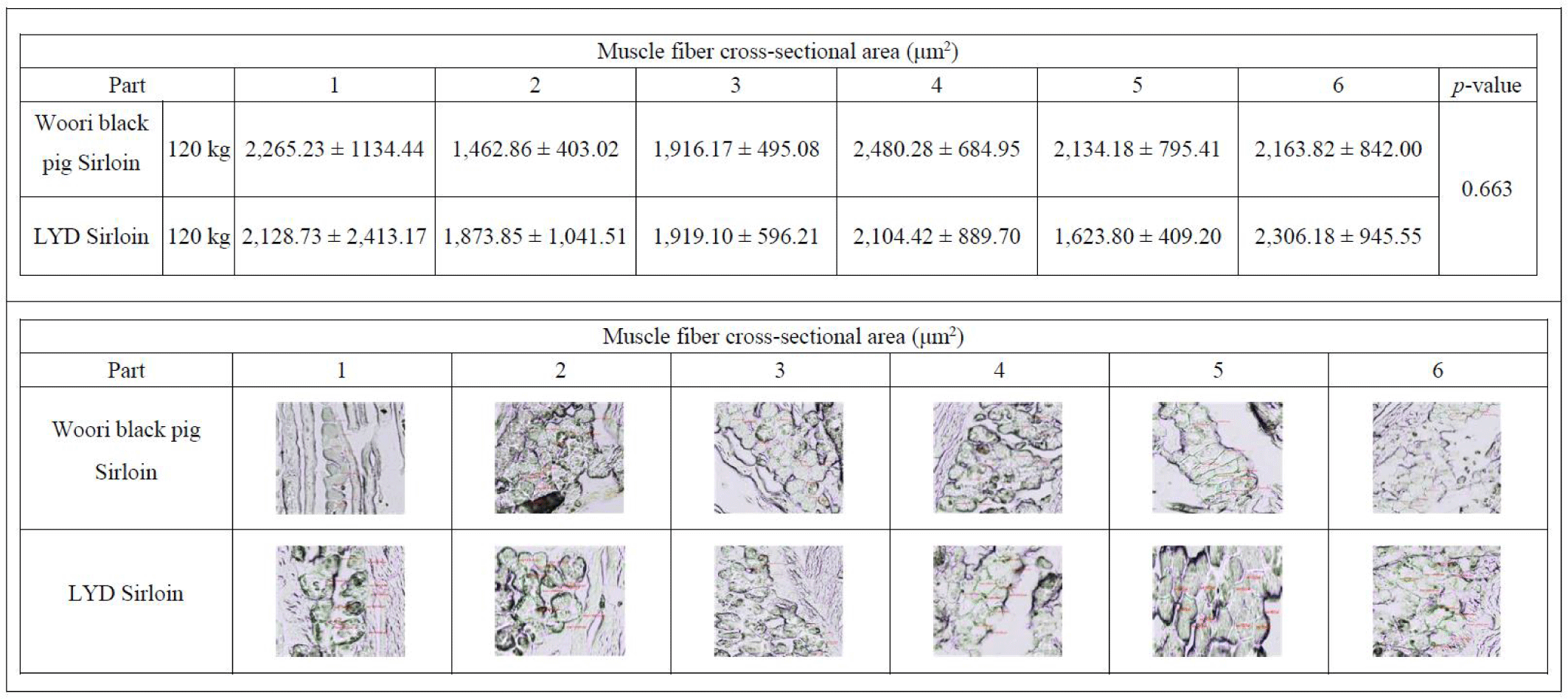
Size of skeletal muscle depends on the size and number of the muscle fiber. In the early stage of growth, the size of the muscle is affected by the number of muscle fibers, and the amount of muscle increases with growing muscle fibers afterward [45]. As 75%–90% of the muscle is composed of muscle fibers [46], total number of fibers and cross-sectional areas play a key role in deciding the muscle weight, juiciness and flavor of the meat [47]. Research conducted by Choi and Oh showed that when there exists little or no difference in types I, IIa, and IIb within the muscle fiber, no significant difference was found in meat tenderness, juiciness, and flavor [48]. Concomitant to these results, there was no difference in the cross-sectional fiber areas in muscles from either WB or LYD (Fig. 7) (p<0.663) indicating that meat quality is not supposed to be different between these breeds.
CONCLUSION
WB is a crossbreed for better meat quality as well as growth efficiency. When comparing growth performance between the two breeds, the final weight of the LYD and WB was 124.37 ± 2.37 kg and 120.63 ± 0.95 kg, respectively. Results of SMSC analysis confirmed that the LYD had more number of SMSC, and PDT was faster than WB. Moreover, as no significant difference was found in sarcomere length, muscle fiber number, and cross-sectional areas, a similar muscle growth rate in the LYD and WB is expected. Gene expression results showed that reduction in myogenesis in SMSC of WB could be induced by down regulation of Pax3 via MSTN (Fig. 8). Expression of adipogenic genes including CEBP-β and FASN, which affect the growth, development, and accumulation of fat, was higher in WB than in LYD. Interestingly, PPAR-γ, which regulates the differentiation of adipose tissue, was expressed lower in WB compare to LYD. Altogether, relatively lower daily weight gain might stem from myostatin activation in WB and results from our current study warrant further investigation of comparative analysis for different pig breeds.