INTRODUCTION
The gut microbiota performs various essential digestive, protective, and metabolic functions for the host’s health [1]. Such benefits include the digestion of complex host-indigestible polysaccharides and endogenous intestinal mucus, pathogen displacement, and synthesis of vitamins [2]. Horses are herbivores whose digestive system has evolved to handle large amounts of a plant-based diet in the large intestine [3–5]. Therefore, horses can obtain energy effectively through fermentation by the microbial activities in their hindgut, mainly in the cecum [1].
Although dietary habits play a major role in regulating the gut microbiota, physical exercise is also considered one of the main environmental factors that might alter the intestinal microbiota [6]. Exercise has many physiological effects, including the improved athletic ability of the bone and muscle, digestion of nutrients, and stimulation of the immune system in humans [7]. The horse study reported that exercise promotes intestinal motility, accelerates the passage rate of intestinal contents, and decreases the contact time between mucosa and pathogens in the intestine [8]. In addition, physical exercise contributes to the production of bile acids and short-chain fatty acids (SCFAs) for energy production in rats, which modifies the gut microbiota [9].
The host’s energy requirement increases during physical activities in humans and animals. Previous studies reported that regular exercise could significantly shift the gut microbial composition, positively affecting energy homeostasis in humans [10]. Exercise can increase the alpha-diversity of the gut microbiota and enhance the gut microbiota-derived SCFAs within athletes [11]. It has been reported that habitual marathon runners had a larger amount of Veillonella, which provided energy sources to the muscle, improving their athletic ability [12]. Overweight women who exercised for six weeks had increased an abundance of Akkermansia that enhanced their metabolic activities while decreasing Proteobacteria that could cause inflammation in the gut [13]. Other studies reported changes in the gut microbiota for Standardbreds and Thoroughbreds, in which the levels of Firmicutes, Bacteroidetes, Proteobacteria, and Spirochaetes phyla increased significantly after training [14].
A horse study reported that fatigue and inadequate recovery cause physical stress, leading to performance decline [15]. Although genetic factors likely play major roles in maintaining the high performance of racehorses, other factors, such as age, conformation, training, diet, and fitness, also affect the racing performance [16]. Several studies have examined the association of the athletic performance with gut microbiota in humans and animals [6,14,17]. The roles of gut microbiota on the racing performance of horses, however, are not entirely understood. The aim of this study was to evaluate the association of the microbial composition and their predicted metabolic activities with the racing performance of Jeju horses and Thoroughbreds in Korea based on the analysis of partial 16S rRNA gene sequence data.
MATERIALS AND METHODS
All animal protocols were approved by the Institutional Animal Care and Use Committee of the Korea Racing Authority (KRA IACUC-2009-AEC-2007). Horse fecal samples were collected from Jeju and Busan-Gyeongnam racecourse in Korea. Forty-nine fecal samples were collected from individual horses: high-performance Jeju horses (HJ, n = 13), low-performance Jeju horses (LJ, n = 17), high-performance Thoroughbreds (HT, n = 9), and low-performance Thoroughbreds (LT, n = 10). Table 1 provides detailed descriptions of the horses used in this study. The Korea Racing Authority (KRA), the regulatory authority for horse racing in South Korea, has their own rating system for racehorses, in which the ability of racehorses is evaluated based on their past racing records. The rating system typically ranges from 0 to 140 with the higher numbers indicating greater racing ability. These scores are calculated based on their past records in races, and these scores are used in the racing industry to determine handicap levels as well as race programs for each horse. With the rating system, horses were classified into 5 different levels. In this study, we used the scores calculated based on scores as of January 2021 and considered classes 1 and 2 as high-performance horses, while classes 4 and 5 as low-performance horses. All horses were selected carefully to minimize the variations in age, body weight (Table 1), diet, training, body condition scoring (BCS), soundness, vaccination, deworming, and medication after undergoing a medical examination (Supplementary Table S1) and checking their medical history and treatment records. Horses were previously acclimated to their racecourses, and no changes in diet, housing, or training conditions were noted for the three months before the study. All horses received roughage, such as alfalfa and timothy, and concentrated feed totally 2.5% to 3% per body weight every day. Jeju horses and Thoroughbreds diets, however, had slightly different diets as shown in Table 2. All horses had access to water ad libitum throughout the study. The fecal samples were collected directly from the rectum to minimize environmental contamination using clean rectal gloves and sterile lubrication (Kruuse, Langeskov, Denmark), as described previously [18]. Each sample was placed in a sealed collection bag and stored at -80°C until DNA extraction.
The fecal DNA was extracted using a PowerFecal DNA extraction kit (Qiagen, Hilden, Germany). The V3 and V4 regions of the partial 16S rRNA gene were amplified by a polymerase chain reaction (PCR) using the 341F and 806R primer sets [19]. Two-step PCR was performed to construct the MiSeq library. Sequencing was performed at Macrogen (Seoul, Korea) according to the manufacturer′s instruction. The sequence data were processed using MOTHUR version 1.45.0 according to the standard operational protocol described online (https://mothur.org/wiki/miseq_sop/) with a minor modification of singleton removal after the pre.cluster subroutine [20]. Silva.nr_v138 was used for alignment, and RDP version 18 was used for the taxonomic classification [21]. The operational taxonomic units (OTUs) were assigned using the opti.clust algorithm with a sequence distance at 0.03 [22]. The PICRUSt2 and MetaCyc database was used to predict the metabolic activities based on the 16S rRNA gene sequences [22]. All sequenced genes were deposited in the NCBI SRA database (accession number; PRJNA817386).
MOTHUR was used to calculate the ecological indices, Chao I and Shannon, for the species richness and diversity, respectively. Non-metric multidimensional scaling (NMDS) was performed and plotted with ellipses at the 95% confidence level using the vegan R package. MOTHUR was used to analyze the molecular variances (AMOVA) to determine the significant differences in the fecal microbiota in the study. Differential abundance analysis was performed using the linear discriminant analysis effect size (LEfSe) [23]. The ALDEx2 R package was used for the OTUs and predicted metabolic activities [24]. A Wilcoxon rank-sum test was applied to compare the ecological indices. The differences were considered significant at p < 0.05.
RESULTS
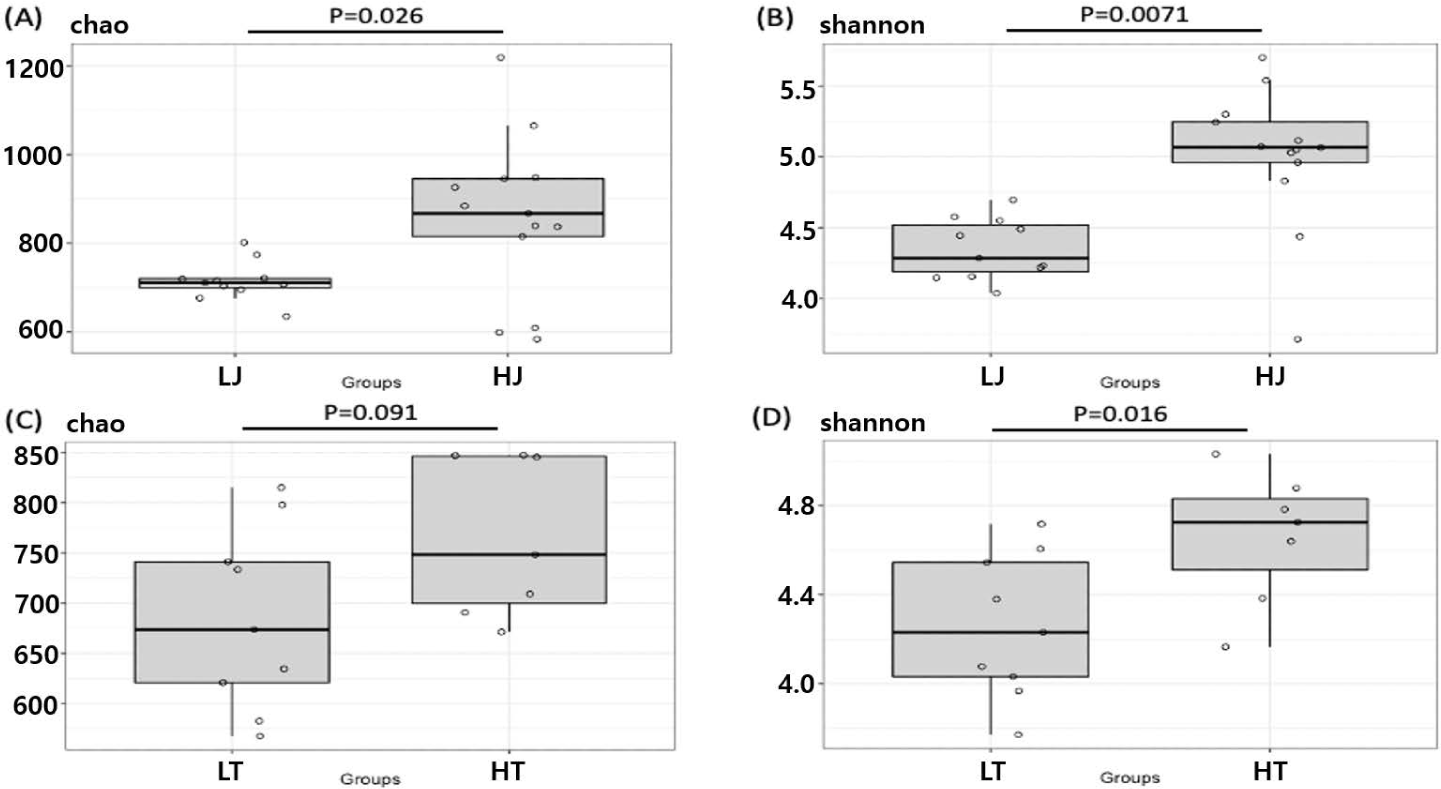
All samples showed a Good’s coverage greater than 98%, suggesting that sequence depth was sufficient to cover most of the species in the samples (Supplementary Fig. S1). The difference in alpha-diversities between the high- and low-performance horse groups was analyzed using the Chao I and Shannon indices for species richness and diversity estimation, respectively (Fig. 1). The species richness of HJ was significantly higher than that of LJ (p < 0.05) (Fig. 1A). HT also had a higher species richness than LT, but the difference was not statistically significant (p = 0.091) (Fig. 1C). The diversity, however, was significantly higher in the high-performance horses for both Jeju horses and Thoroughbreds (p < 0.05) (Figs. 1B and 1D).
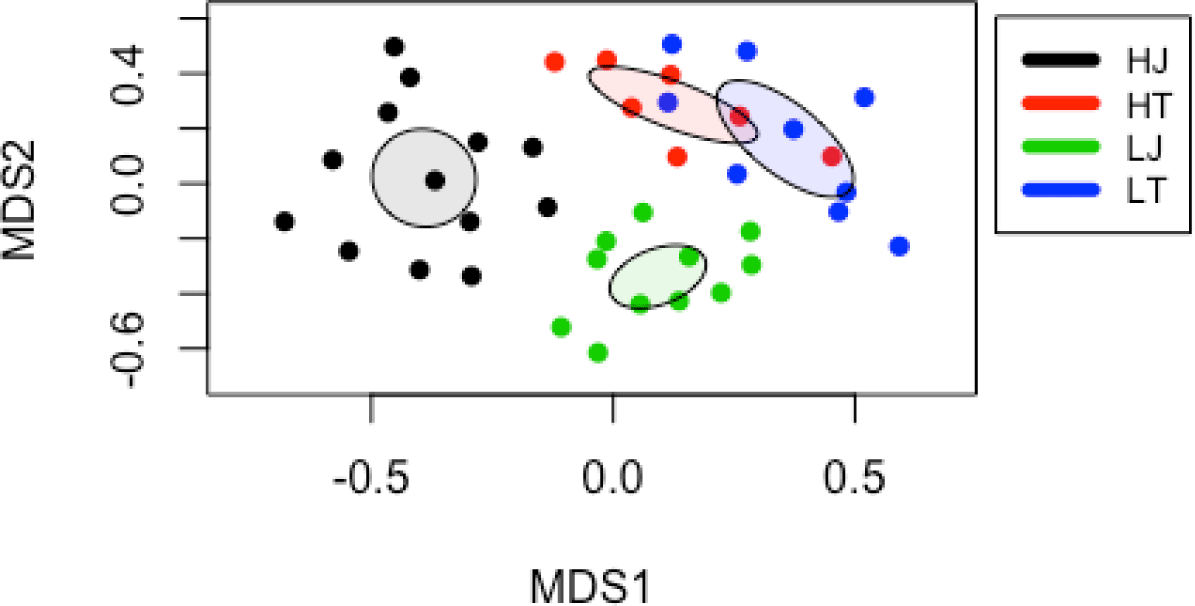
Based on NMDS analysis, the beta-diversity showed that the fecal microbiota of the high-performance horse groups was significantly different from each counterpart (p < 0.05) (Fig. 2), indicating that the gut microbiota affects the racing performance. Although the distance of gut microbiota between HT and LT groups was closer than that of HJ and LJ groups in NMDS analysis, the results from AMOVA suggested that HT is significantly different from LT microbiota (Supplementary Table S2) (p < 0.05).
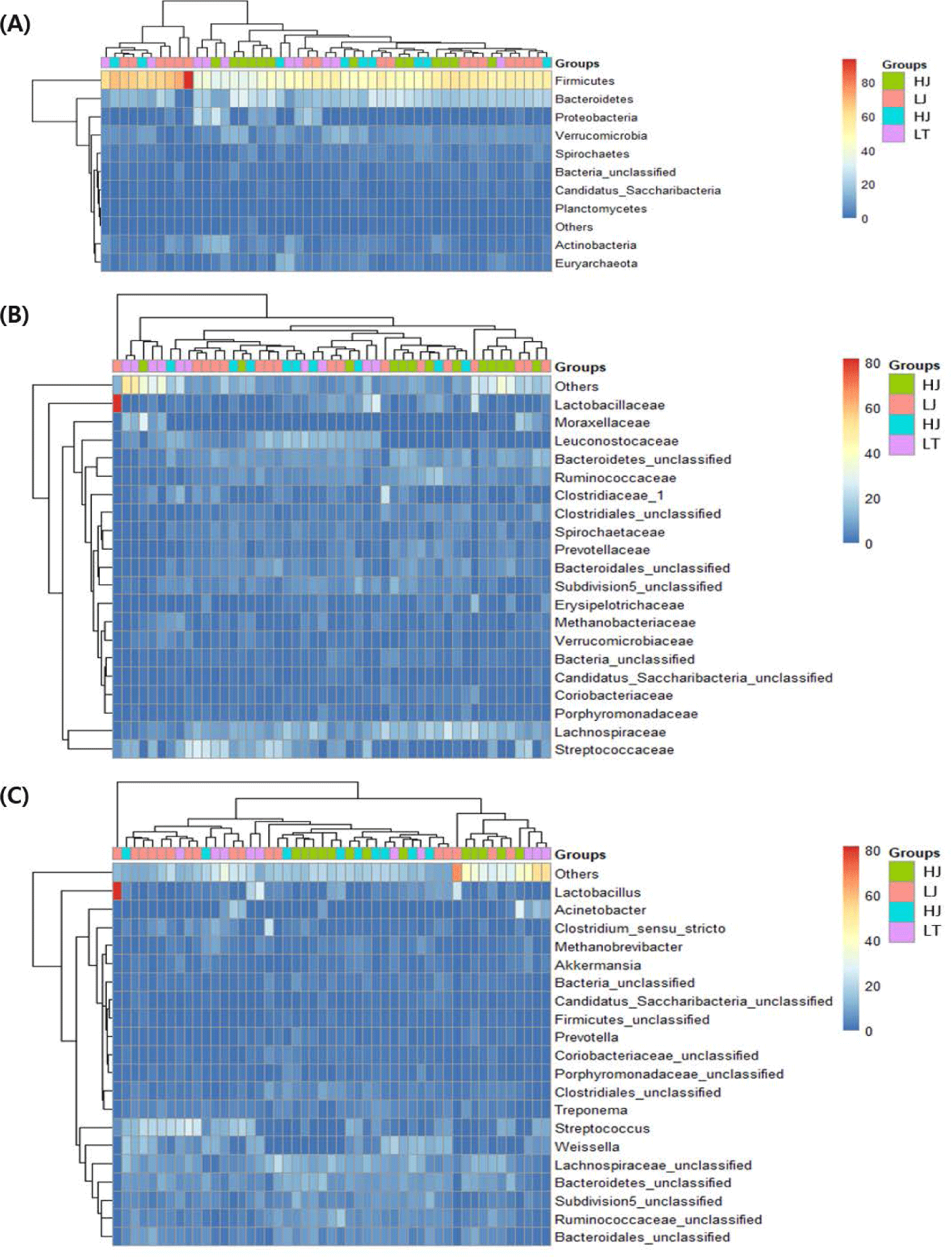
Comparisons of the fecal microbial communities were performed at the bacterial phylum, family, and genus levels (Fig. 3). Firmicutes and Bacteroidetes were the predominant phyla, followed by Proteobacteria and Verrucomicrobia. Among the Jeju horses, Firmicutes were more abundant in LJ, while Actinobacteria was more in HJ (Supplementary Fig. S2A). In contrast, Actinobacteria were more abundant in LT, and Spirochaetes were more abundant in HT (Supplementary Fig. S2B). At the family level, Ruminococcaceae was more abundant in both high-performance horses (p < 0.05) (Supplementary Figs. S2C and S2D).
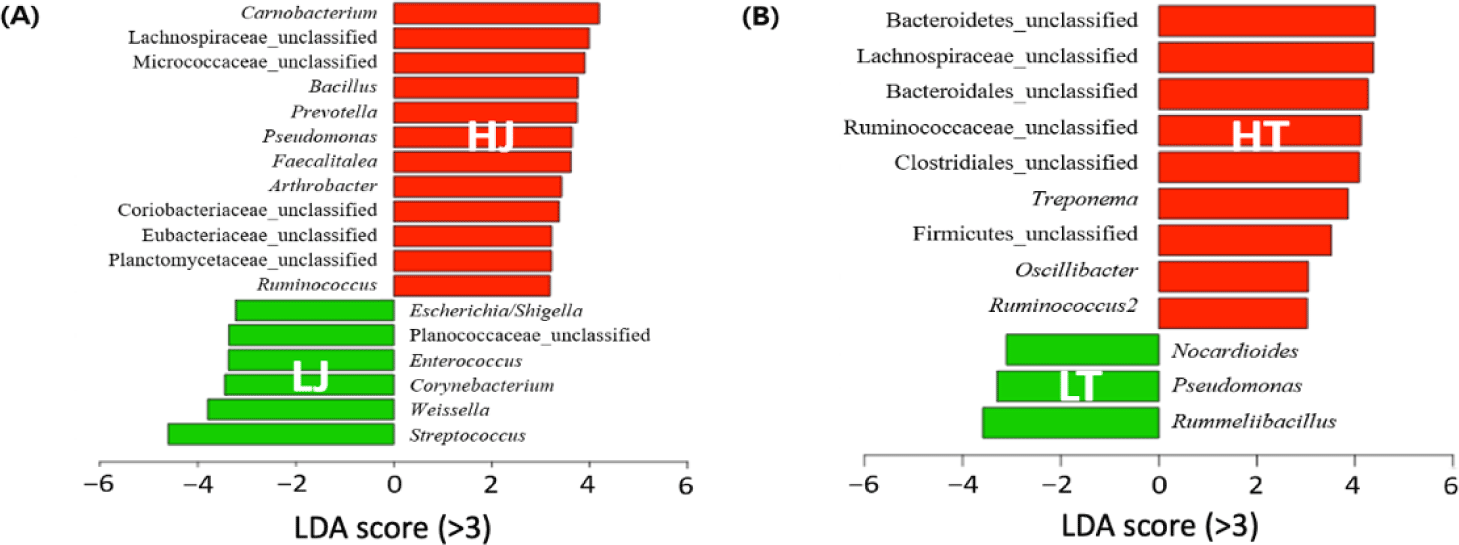
The differentially abundant genera in all groups were identified by LEfSe (Fig. 4). Significant differences were observed between the fecal microbiota of high- and low-performance horses in both breeds (p < 0.05). High performance horse groups showed a significantly higher abundance of fiber fermenting bacteria compared to low-performance horse groups (p < 0.05). Specifically, the HJ group exhibited a greater abundance of Lachnospiraceae_unclassified, Prevotella, and Ruminococcus, while the HT group had a higher abundance of Lachnospiraceae_unclassified, Ruminococcaceae_unclassified, Oscillibacter, and Ruminococcus2 (p < 0.05). By contrast, pathogenic species were found to be more abundant in the low-performance group. Escherichia/Shigella, Enterococcus, and Streptococcus were more abundant in the LJ group, while Pseudomonas was more abundant in the LT group (p < 0.05). Treponema, some species of which are known as human pathogenic bacteria, was more abundant in HT [25].
Tables 3 and 4 list the significantly enriched metabolic activities of the fecal microbiota among the high-performance horse groups compared to those of low-performance horse groups (p < 0.05). Among the HJ group, metabolites related to polyamine syntheses, such as L-methionine salvage cycle III (PWY-7527) and norspermidine biosynthesis (PWY-6562), plant-derived fiber digestion (i.e., HYDROXYPHENYLACETATE-DEGRADATION-PWY), and methanol oxidation (PWY-7616) were enriched. The HT group, however, was enriched with the metabolic activities involved in plant-derived fiber digestion, such as rhamnose (PHAMCAT-PWY) and mannan (PWY-7456), and the production of SCFAs and vitamins (e.g., demethylmenaquinol-6 [PWY-7373]).
On the other hand, there were five metabolic pathways (i.e., PWY-6629, PWY-6165, ORNDEG-PWY, ARGDEG-PWY, and ORNARGDEG-PWY) enriched among the low-performance horse groups in both LJ and LT (Tables S3 and S4). The metabolites involved in these metabolic pathways included L-tryptophan, chorismate, 4-aminobutanoate (GABA), and succinate.
DISCUSSION
Considering that the intestinal microbiota is sensitive to many factors, including the environment, diet, and age, Physical exercise is also associated with the positive modulation of intestinal microbial diversity. The current study examined the association of the gut microbiota on the racing performance of horses.
A comparison of the alpha-diversity revealed a higher species diversity in high-performance horse groups than in low-performance horse groups. In addition, significantly different beta-diversity was observed among the groups (p < 0.05). Exercise increases the diversity of human gut microbiota, and the mode and intensity of exercise affect the degree of changes in gut microbiota [26–28]. Moreover, Liu et al. reported that muscle phenotypes can be directly affected by altering the gut microbiota [29]. Together, based on previous studies [26–29], it can be inferred that the racing performance of Jeju horses and Thoroughbreds in Korea is likely affected by the composition of the intestinal microbiota.
The normal horse gut microbiota comprises two major phyla, Firmicutes and Bacteroidetes, and to a lesser extent, Verrucomicrobia, Euryachaeota, and Spirochaetes [1,18]. In the present study, a higher abundance of Actinobacteria was observed in HJ than HT. Because Jeju horses and Thoroughbreds have different baseline gut microbiota [18], the effects of exercise on the gut microbiota may differ. High-intensity exercise that exceeds an individual’s ability may also adversely affect the gut microbiota [30].
The high-performance horse groups had significantly different compositions of fecal microbiota from their counterparts (p < 0.05). Physical exercise modified various phyla with an increase in Bacteroidetes and a decrease in Firmicutes regardless of diet [17]. Since animal and human studies have shown that the F/B ratio is a relevant marker of obesity, the ratio may also indicate variations in capacities of fat storage, energy collection from nutrients, and energy expenditure [31]. In this study, the F/B ratios did not show a significant difference, but higher Firmicutes (p < 0.05) were observed in the LJ group than in the HJ group.
Fiber fermenting bacteria were found to be significantly more abundant in the high-performance horse groups than in the low-performance horse groups (p < 0.05). By contrast, pathogenic species were found to be more abundant in the low-performance group (p < 0.05). Several commensal fiber-digesting bacteria, such as Lachnospiraceae_unclassified, Ruminococcaceae_unclassified, Ruminococcus, Ruminococcus2, Prevotella, and Oscillibacter, were more abundant in the high-performance horse groups than the low-performance horse groups [32–34]. Lachnospiraceae assists in the digestion of indigestible polysaccharides in humans and horses [32]. Many of the species belonging to the family Ruminococcaceae also breaks down the fiber effectively and produces butyrate, which is one of the major SCFAs found in the intestines of herbivores [33]. Prevotellaceae is abundant in horses living on pasture and degrades the proteins and carbohydrates [34]. Lachnospiraceae, Ruminococcaceae, and Oscillibacter promote fermentation and produce SCFAs as energy sources in horses and other animals [32,33]. Faecalitalea, whose abundance was higher in HJ, may produce butyrate and polyphenols with antioxidant activities [35], thereby benefiting intestinal health and fatigue recovery [36].
Coriobacteriaceae_unclassified was higher in HJ than LJ in the present study. Coriobacteriaceae family has been reported to increase in the human gut after physical exercise, such as long-distance running [36]. This bacterial family is involved in converting polyphenols to bioactive derivatives and in the metabolism of bile salts and aldosterone. The metabolite of aldosterone holds important functions, such as fuel and energy storage and membrane stability [37]. Therefore, the Coriobacteriaceae family was also a potential biomarker linking exercise with health improvement [37]. Thus, having a more abundant Coriobacteriaceae family, Faecalitalea, that help generate energy is seemed to influence high-performance horse groups to achieve good race records.
Pseudomonas, Escherichia/Shigella, Enterococcus, and Streptococcus, were more abundant in the low-performance horse groups. Some species of Pseudomonas causes glanders, which is a contagious zoonotic infectious disease in humans and horses [38]. Some species of Escherichia/Shigella and Enterococcus cause colitis [39]. Some species of Streptococcus causes strangles, meningitis, and colitis in horses [39]. The higher abundance of Treponema, a pathogenic bacterium, in horses that underwent training is consistent with previous studies [25]. In this study, statistic comparison did not show significant differences neither for age nor body weight (Table 1), thus our study indicated the race performance as a single feature associated with gut microbiota.
The high-performance Jeju horse group showed enriched metabolisms related to polyamine biosynthesis, while the HT showed enriched SCFA and vitamin production. Polyamines produced in the gut have a positive effect in regulating the intestinal permeability by controlling intestinal tight-junctions [40], while SCFA provides diverse beneficial health effects, including energy to epithelial cells and regulating immunity. Vitamin K produced in the gut prevents blood coagulation [41]. Together, these metabolisms improve intestinal health. Moreover, enriched metabolisms of methanol oxidation were observed in HJ, which were previously suggested as a marker of healthy horses [42].
On the other hand, metabolites involved in tryptophan and succinate syntheses were enriched among low-performance horses. Tryptophan is an ingredient used as calmatives for fearful or excitable horses [43]. Farris et al. reported that the horses given tryptophan showed a tendency to use less muscle glycogen during exercise [44]. In addition, tryptophan plays a role as a substrate for the synthesis of serotonin. The serotonin activity is associated with fatigue and increases during prolonged exercise [45]. To horses, the amount of serotonin was reported to be negatively correlated with dominance [46], suggesting that horses with a higher amount of serotonin may be less likely to win races [47,48]. Moreover, the amount of serotonin has been associated with fatigue in athletic horses [49]. Succinate, however, is an intermediate of the tricarboxylic acid cycle and is produced in large amounts during the bacterial fermentation of dietary fiber [50]. On the other hand, it was reported that elevated succinate levels in fecal microbiota were associated with microbial disturbances (dysbiosis) [50], which could be related to the abundance of potentially pathogenic bacteria.
As in previous studies [26–29,51], despite the results revealing the significant relationship between gut microbiota metabolism and racing performance (p < 0.05), there were some limitations in analyzing the metabolic activities because PICRUSt may show less accuracy in predicting the metabolic activities in non-human fecal samples. Further investigation should include a metabolomics approach to understand the associations of gut bacteria-derived metabolites and athletic performance in horses.
In conclusion, this study examined the association between gut microbiota and racing performance in Jeju horses and Thoroughbreds. The high-performance horse groups have a more balanced gut microbiota composition than the low-performance horse groups. The high-performance horse group showed higher diversity with beneficial bacteria and indicated some beneficial gut microbiota-derived metabolic activities, such as the production of polyamines and SCFAs. The low-performance horse groups, however, showed more bacteria, many species of which include pathogens, and non-beneficial metabolic activities for athletic horses.
















