RESEARCH ARTICLE
Complete genome sequence of Escherichia coli K_EC180, a bacterium producing shiga-like toxin isolated from swine feces
Hyeri Kim1,#
,
Jae Hyoung Cho1,#
,
Jin Ho Cho2,#
,
Minho Song3,#
,
Hakdong Shin4
,
Sheena Kim1
,
Eun Sol Kim1
,
Hyeun Bum Kim1,*
,
Ju-Hoon Lee5,*
Author Information & Copyright ▼
1Department of Animal Resources Science, Dankook University, Cheonan 31116, Korea
2Division of Food and Animal Science, Chungbuk National University, Cheongju 28644, Korea
3Division of Animal and Dairy Science, Chungnam National University, Daejeon 34134, Korea
4Department of Food Science and Biotechnology, College of Life Science, Sejong University, Seoul 05006, Korea
5Department of Food Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul 08826, Korea
*Corresponding author: Hyeun Bum Kim, Department of Animal Resources Science, Dankook University, Cheonan 31116, Korea. Tel: +82-41-550-3653, E-mail:
hbkim@dankook.ac.kr
*Corresponding author: Ju-Hoon Lee, Department of Food Animal Biotechnology, Department of Agricultural Biotechnology, Center for Food and Bioconvergence, Seoul National University, Seoul 08826, Korea. Tel: +82-2-880-4854, E-mail:
juhlee@snu.ac.kr
#These authors contributed equally to this work.
© Copyright 2021 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Oct 22, 2020; Revised: Jan 20, 2021; Accepted: Jan 21, 2021
Published Online: Mar 31, 2021
Abstract
Escherichia coli normally colonizes the lower intestine of animals and humans, but some serotypes are foodborne pathogens. The Escherichia coli K_EC180 was isolated from swine feces that were collected from a weaner pig. In this genome announcement, E. coli K_EC180 was sequenced using PacBio RS II and Illumina NextSeq 500 platforms. The complete chromosome of E. coli K_EC180 is composed of one circular chromosome (5,017,281 bp) with 50.4% of guanine + cytosine (G + C) content, 4,935 of coding sequence (CDS), 88 of tRNA, and 22 of rRNA genes. The complete genome of E. coli K_EC180 contains the toxin genes such as shiga-like toxins (stxA and stxB).
Keywords: Escherichia coli K_EC180; Swine feces; Whole genome sequencing; Shiga-like toxin
INTRODUCTION
Escherichia coli is a facultative anaerobic bacterium which is commonly spread on biosphere. E. coli normally colonizes the lower intestine of animals and humans (1). However, Some of the serotypes such as Enterohemorrhagic E. coli (EHEC), Enterotoxigenic E. coli (ETEC), Enteropathogenic E. coli (EPEC) and Shiga toxin-producing E. coli (STEC) can cause foodborne illnesses in people.
E. coli K_EC180 was isolated from swine feces that were collected from a livestock farm in Haenam-gun, Jeollanam-do, Korea. E. coli K_EC180 was streaked to Luria-Bertani (LB) agar and incubated at 37°C for 24 h. The suspected colony in LB agar was inoculated into LB broth and incubated at 37°C for 24 h. To analyze the complete genome, the E. coli K_EC180 genome was sequenced by PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) at Insilicogen (Yongin, Korea) and Illumina NextSeq 500 (Illumina, San Diego, CA, USA) platform at LabGenomics (Seongnam, Korea). The genomic DNA of E. coli K_EC180 for PacBio and Illumina sequencing was extracted using the MagAttract HMW DNA Kit (QIAGEN), and NucleoSpin® Microbial DNA kit (TAKARA) according to the manufacturer’s instructions. Library preparation was conducted using SMRTbell™ Template Prep Kit 1.0 for Pacbio (Pacific Biosciences) and TruSeq DNA Sample Preparation Kit for Illumina (Illumina) according to the manufacturer’s instructions. PacBio sequencing yielded 1,131,537,370 base pairs and 145,423 long reads after filtering, and 9,199,306 paired-end reads with 1,389,095,206 bp were obtained with Illumina sequencing. De novo assembly was conducted using the hierarchical genome assembly process (HGAP v2.3.0) workflow (Chin et al., 2013) and polished using Quiver. Subsequently, Illumina NextSeq reads were aligned to the PacBio RSII assembly using Burrows-Wheeler Aligner (BWA)-MEM v0.7.17-r1188, and the errors were corrected by using Pilon version 1.23 (2, 3). The quality of genome assembly and the validaty of the final genome were assessed using Quality Assessment Tool for Genome Assemblies (QUAST) v5.0.2 and Benchmarking Universal Single-Copy Orthologs (BUSCO) v3.0.2 (4, 5). Open reading frames (ORFs) and RNA genes of E. coli K_EC180 were predicted and functionally annotated through rapid prokaryotic genome annotation (PROKKA) v1.14.5 (6) and Rapid Annotation using Subsystem Technology (RAST) v2.0 (7). The functional categorization and classification of all predicted ORFs were conducted using the RAST server-based SEED viewer and Clusters of Orthologous Groups (COG) – based EggNOG. The putative virulence factors and Antimicrobial resistance were described using BLAST according to the Virulence Factor Database (VFDB) (8). The whole genome of E. coli K_EC180 is composed of one circular chromosome (5,017,281 bp) with 50.4% of G+C content, 4,935 of coding sequence (CDS), 88 of tRNA, and 22 of rRNA genes.
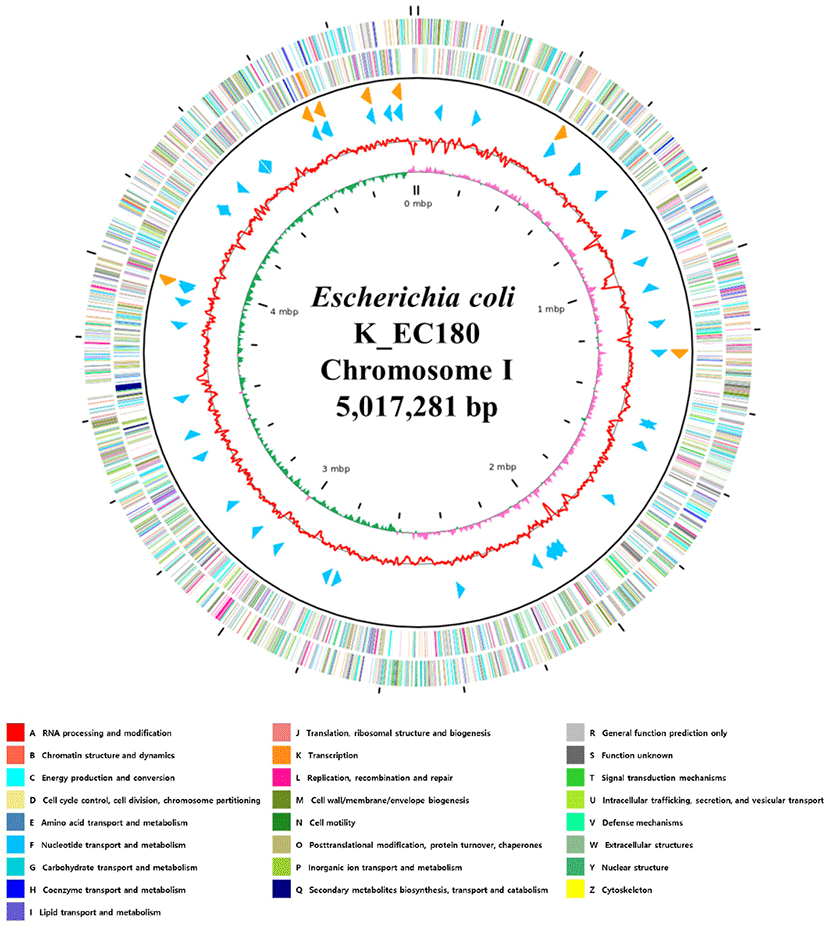
The complete genome of E. coli K_EC180 contains the toxin genes encoding shiga-like toxin (stx2e subunit A and stx2e subunit B), which may cause diseases in humans by damaging small blood vessels in places such as the digestive tract, kidneys and central nervous system (9, 10). E. coli K-EC180 also possessed essC, escV, escR, escS, escV, and escJ genes which involved in a type III secretion system. In addition, there were fim (A to H) genes encoding Type I fimbriae. We summarized the general properties of the E. coli K_EC180’s complete genome in the Fig. 1 and Table 1.
Fig. 1.
Genome map of Escherichia coli K_EC180.
The outer circle denotes the locations of all annotated ORFs, and the inner circle with the red denotes GC content. Pink, and green peaks denote GC skew. The orange arrows denote rRNAs, and the sky blue arrows denote the tRNA operons. All annotated ORFs are colored differently based on the COG assignments. ORFs, open reading frames; G, guanine; C, cytosine; COG, clusters of orthologous groups.
Download Original Figure
Table 1.
Genome features of Escherichia coli K_EC180
| Property |
Term |
| Libraries used |
PacBio SMRTbell™ library
TruSeq DNA Sample Preparation Kit |
| Sequencing platforms |
PacBio RS II sequencer
Illumina NextSeq 500 |
| Assemblers |
PacBio SMRT analysis v2.3.0 HGAP.3 |
| Annotation method |
PROKKA v1.14.5 and RAST v2.0 |
| Average genome coverage |
100× |
| Chromosome length (bp) |
5,017,557 bp |
| No. of contigs |
1 |
| Guanine + cytosine (G + C) content (%) |
50.4 |
| Protein–coding genes (CDSs) |
4,935 |
| rRNA genes |
22 |
| tRNA genes |
88 |
| Plasmids |
0 |
| Genbank Accession No. |
CP062203 |
Download Excel Table
DATA AVAILABILITY
The complete genome sequences of E. coli K_EC180 were deposited in GeneBank under the accession numbers CP062203. The BioSample accession number is SAMN16277032, and BioProject accession number is PRJNA666028.
Acknowledgements
We thank Mo Re Kim (Brandeis University, MA, USA) for the English grammar corrections.
Availability of data and material
Ethics approval and consent to participate