ANNOUNCEMENT
Limosilactobacillus fermentum (formerly named Lactobacillus fermentum JN2019) is commonly found in fermented food products and is generally considered safe [1]. L. fermentum has been regularly used for acid-producing starter cultures and acts as a food preservative [2]. In addition, JN2019 increases the bioavailability of curcumin, the active component of turmeric, while reducing cytotoxicity through fermentation [3]. In the present study, the JN2019 genome was sequenced to explore its genetic characteristics.
JN2019 was isolated from local fermented kimchi in Korea and grown in de Man-Rogosa-Sharpe (MRS) medium (Merck, Darmstadt, Germany). Genomic DNA (gDNA) was extracted with DNeasy Ultraclean microbial kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The gDNA was sequenced using single molecular real-time (SMRT) portal (v.2.3) with the PacBio RS II system (Pacific Biosciences, Menlo Park, CA, USA). A total of 43,479,132 reads (6,565,348,932 total bases) were generated using SMRT sequencing. Gene neighborhood analysis illustrating the closest genome to JN2019 was L. fermentum strain DR9 (98.26%), followed by strain FTDC 8312 (85.60%). Although pronounced similarity to strain DR9 was observed, the value fell below the species recognition threshold of 98.6% [4], clarifying the identity of JN2019 as L. fermentum. The genome sequences were annotated by the NCBI Prokaryotic Genomes Annotation Pipeline.
The complete genome of JN2019 consists of two contigs within the 2.3 Mb genome (G + C content of 50.5%), a single chromosome of 2.3 Mb with a G + C content of 50.6% and a 30 kb plasmid with a G + C content of 40.8% (Table 1). The 2.3 Mb genome corresponds to 2,359 genes, 2,077 proteins, 15 rRNAs, 58 tRNAs, and 3 other RNAs. These 2,359 genes are specifically clustered into 26 Clusters of Orthologous Groups of proteins-based functional categories (Fig. 1).
| Chromosome | Plasmid | |
|---|---|---|
| Genome size (bp) | 2,298,221 | 29,243 |
| G + C content (%) | 50.6 | 40.8 |
| Gene | 2,359 | 34 |
| Pseudogene | 206 | 6 |
| Protein | 2,077 | 28 |
| rRNA | 15 | - |
| tRNA | 58 | - |
| Other RNA | 3 | - |
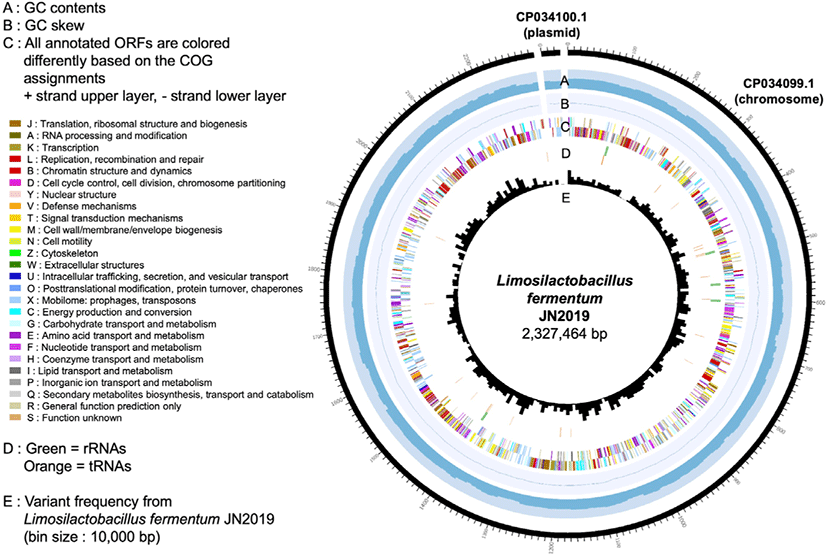
The genome information of JN2019 provides fundamental knowledge to inform discoveries of its beneficial properties and industrial applications. The complete genome sequence of JN2019 is available from NCBI/GenBank under BioSample accession number SAMN10417155 or directly via the assembly accession number CP034099.1 (chromosome) and CP034100.1 (plasmid).
















