INTRODUCTION
Most of the genes refer to a region of a DNA sequence that functions related to animal traits or diseases. Therefore, gene expression profiling has been used to analyze biological functions [1], and analysis has been conducted by reading RNA sequences from the transcription process of DNA. However, coding RNAs that are translated into protein accounts for only about 4% of RNA, and the fact that non-coding RNAs (ncRNAs) existing in a vast region, which were treated with no role in the early days, are involved in gene regulation in various ways are being investigated [2].
Among them, long non-coding RNA (lncRNA), unlike mRNA, is not translated into a protein, despite its similar sequence structure [3]. In a small number of investigations involving animals, plants, and humans, it has been revealed that lncRNA functions in certain diseases or specific environments. It turns out that lncRNAs, previously considered to have no role, play many significant roles, the most important of which is to regulate mRNA expression [4,5]. LncRNAs regulate gene expression in a variety of ways at epigenetic, chromatin remodeling, transcriptional, and translational levels [6]. With the development of Next Generation Suquencing, lncRNA identification has been performed in humans and plants but also various species of animals. As the studies progressed, it was found that lncRNA had longitudinal, tissue-specific, and environmental-specific properties, so various case studies began to progress in various animals [4]. Prior studies and database construction are insufficient in other animals compared to humans and mice, so efforts are underway to continuously discover lncRNAs with essential functions and to be studied in many livestock animal samples [7–9]. However, even after some time since the importance of lncRNA emerged, many lncRNA transcripts have not been identified in livestock animals, or the functions of lncRNAs have not been identified properly. Therefore, lncRNA research is expected to be actively conducted for higher-dimensional bioinformatic analyses and multi-omics integration (MOI).
RNA is a polymeric genetic material that plays a vital role in various life phenomena, including control of gene expression [10,11]. Unlike DNA, RNA is not a pair of double strands but a single-stranded molecule with a short chain of nucleotides [12]. Notably, RNA can be divided into two main categories: messenger RNA (mRNA), which is coded as protein, and ncRNA, which is not coded [13].
For DNA genetic information to be expressed as a protein, DNA must first be transcribed into RNA, and this RNA transcribed to be translated into protein is called mRNA [14]. With the development of sequencing technology, it became possible to examine the transcriptome region and to identify genes representing functions. Therefore, studies on mRNA expression levels under various conditions and bioinformation analysis-related studies using these results are being actively pursued [15–17]. In humans, mRNA is mainly used for pharmaceutical and vaccine development by enhancing the understanding of the immune system [18–24]. Additionally, various studies in mice are being conducted for use in humans because mice are also very similar in their genes to humans [25–30]. Furthermore, it is widely used for various trait studies in livestock animals. Many mRNA-related studies have been conducted mainly for the analysis of animal production traits [31–39] and quality traits [40–44], and they are also used for research in a wide range of areas, such as milk production [45–48], egg production [49–51], nutrients [52–54], stress [55–60], disease [61–64], and reproductive traits [65–71].
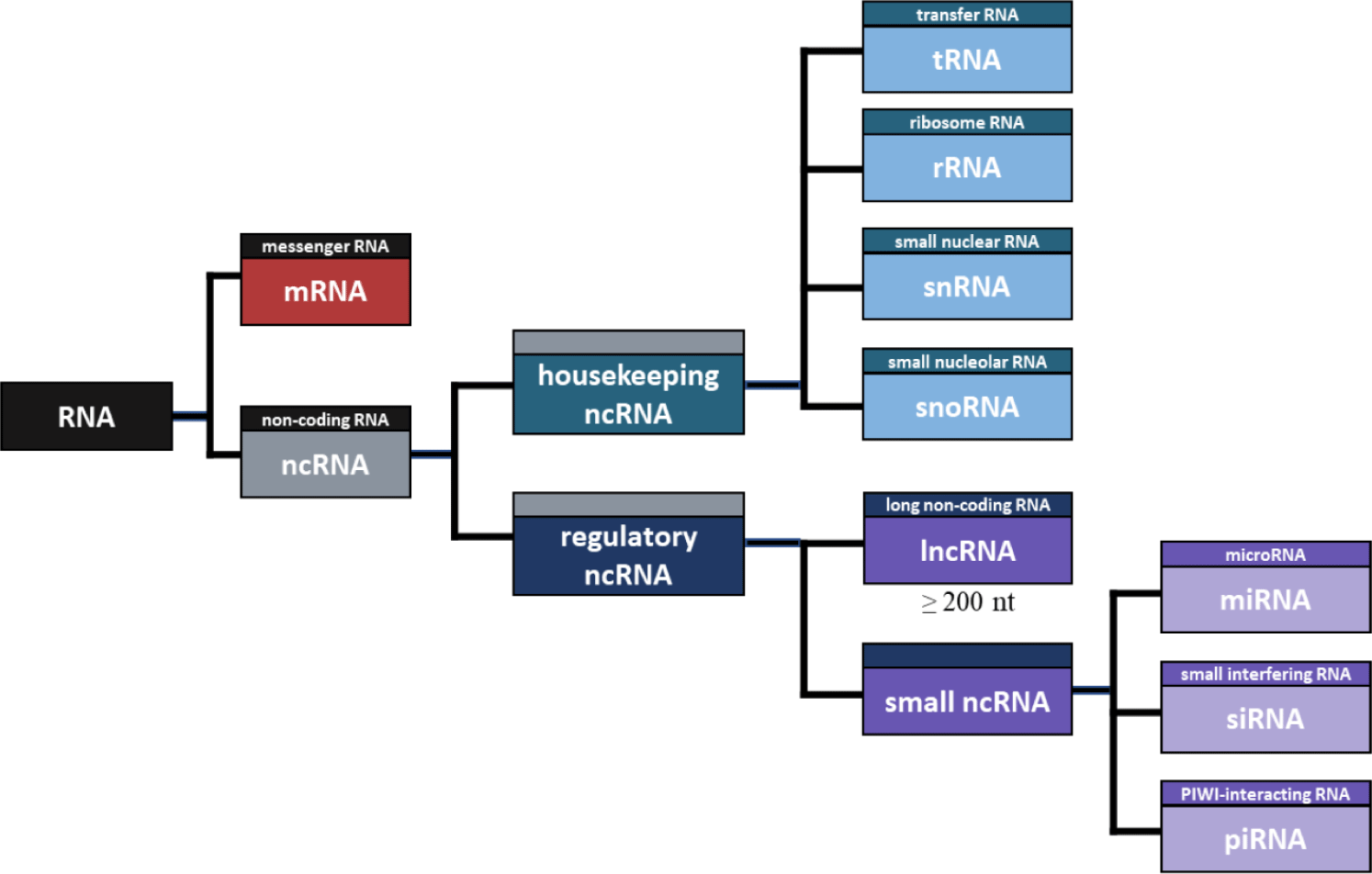
NcRNA refers to RNA that is not translated into a protein [72], and there are many different types of ncRNA. First, ncRNA can be divided into housekeeping and regulatory ncRNA (Fig. 1). Housekeeping ncRNAs include transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA), and small nuclear RNA (snoRNA), while regulatory ncRNAs include microRNAs (miRNA), small interfering RNA (siRNA), piwi-interacting RNA (piRNA), and lncRNA [73]. Housekeeping ncRNAs are essentially expressed and mainly involved in rRNA modification RNA splicing control [74]. tRNA has a complementary anticodon in protein synthesis, which carries the amino acid to mRNA [75], and rRNA is an RNA that plays a structural role in ribosome formation and contributes to enzyme activity for protein synthesis [76]. Additionally, SnRNA binds with other proteins to form snRNPs, and plays a role in recognizing introns in the splicing process [77]. SnoRNA is primarily responsible for chemical transformation, such as rRNA and tRNA [78]. Notably, the main difference between the two RNAs is that snRNA influences the alternative splicing of pre-mRNA molecules to determine which sequence should be translated into proteins. In contrast, snoRNA participates in tRNA, rRNA, and mRNA editing and genome imprinting [79].
Regulatory ncRNAs can be divided into small RNAs and lncRNAs according to the length of RNA. The small ncRNA includes miRNA, siRNA, piRNA, and the like. MiRNA is an ncRNA composed of about 22 nt and functions in RNA silence and regulation of gene expression after transcription [80]. Likewise, SiRNA is an ncRNA composed of approximately 23 nt, which is involved in RNA interference and interferes with gene expression by inhibiting the production of specific proteins [81]. The main difference between the two RNAs is that miRNA regulates the expression of several mRNAs, and siRNA inhibits the expression of specific target mRNAs [82]. Furthermore, PiRNA consists of about 30 nt, which induces PIWI proteins to cleave the target RNA, promote heterochromatin assembly, methylate DNA, and regulate gene expression [83].
Among regulatory ncRNAs, RNA molecules greater than 200 nt in length are defined as lncRNA [84]. Although lncRNAs are very similar in structure to mRNAs, they are not translated into proteins and regulate gene expression through various bases, including epigenetic modification [3]. The various lists of annotated lncRNAs based on resemblance to protein-coding mRNAs account for only 0.05%–1.12% of cellular RNA, while functional intronic RNAs could constitute as much as 16% [85].
The extensive sequences that do not encode proteins (i.e., the majority of the vast regions of intronic and intergenic sequences) have been regarded as accumulated evolutionary remains arising from the early assembly of genes and/or the insertion of mobile genetic elements. However, as the aforementioned regulatory ncRNAs show, most of these supposedly inert sequences are transcribed and widely employed for gene regulation in cis and trans [86].
LONG NON-CODING RNA
Although the structure of the lncRNA seems similar to that of the mRNA, lncRNA is not coded and exists as a ncRNA rather than mRNA.
LncRNAs mostly have a cap at the 5´ end and a poly(A) tail at the 3´ end, presumed to be transcribed similarly to mRNAs [87]. LncRNAs are transcribed by RNA Polymerase II (Pol II) and RNA Polymerase III at several loci of the genome, most of which are transcribed by Pol II [88]. Due to the lncRNA having a weak internal splicing signal and having a long distance between the 3’ splice site and the junction, the lncRNA is spliced more inefficiently than the mRNA [89–91]. The nuclear position and fate of lncRNAs appear to be coordinated by various causes, ranging from transcription to nuclear export via sequence motifs in cis and factors in trans [4,92]. Since the arrangement and size of lncRNAs are diverse, it is not well known precisely what biogenesis pathways they are treated. It is also challenging to understand whether ribosome-related lncRNAs are involved by ribosomes for translation, so further research is needed [4].
Unlike small ncRNAs such as siRNAs, miRNAs, and piRNAs, lncRNAs are relatively long and therefore have poorly conserved properties [93]. Compared to mRNA, lncRNA has a shorter transcript length and a smaller number of exons on average, and many studies have demonstrated these characteristics [94–96]. Furthermore, lncRNA has a shorter open reading frame (ORF) length than mRNA and a relatively low expression level [89,97,98].
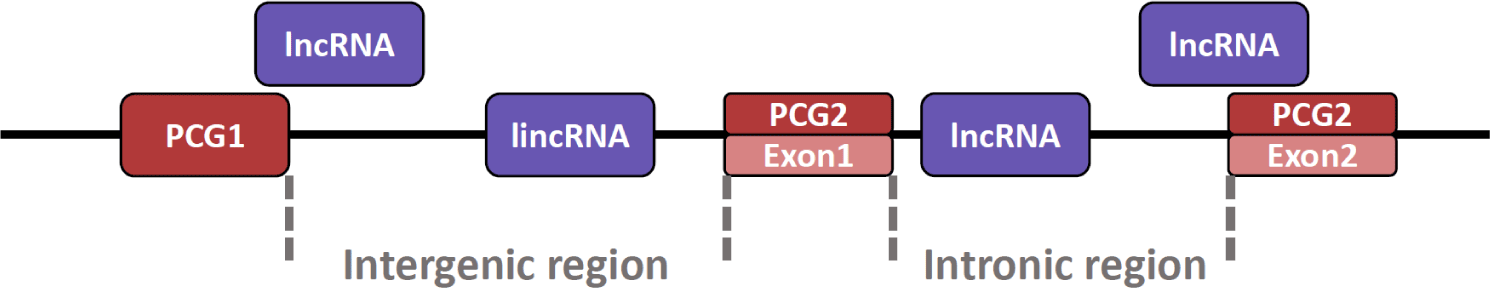
Also, lncRNA can exist at various locations in the genome (Fig. 2) [99]. The lncRNA can be present in the intron region between exon and exon and in the intergenic region between the protein-coding gene (PCG) and PCG [100]. In particular, lncRNAs present in the intergenic region are named long intergenic non-coding RNA (lincRNA). Additionally, because lincRNA does not overlap with PCG domains or other small RNA genes, it is relatively easy to conduct research such as the structure and function of lincRNA [101]. Although lincRNAs are similar in many respects to lncRNAs, they are somewhat longer than lncRNAs and are characterized by their presence in mammalian nuclei [101,102]. Furthermore, lncRNA can also exist in an exonic region where the lncRNA transcript overlaps the exon portion of the PCG [103]. Notably, there are also antisense lncRNAs characterized by transcription from opposite strands of PCG [104], which regulate the expression of their endogenous sense genes [105].
LncRNA has well-known tissue-specific, species-specific, and conditional-specific tendencies. Even the same individual can express lncRNA differently depending on what kind of tissue it is, and even the same tissue can express differently depending on the species [96,106–110]. Moreso, the tissue-specific characteristics of lncRNA demonstrated higher results even when compared to mRNA through the tissue specificity index calculated numerically in mammals [96,111].
Most animals, except humans and mice, do not yet have a well-established lncRNA database, so the process of identifying novel lncRNA identification for lncRNA analysis should be conducted. Therefore, a new Gene transfer format (GTF) file is needed to find the novel lncRNA instead of the reference gtf file of the animal containing only the information of previously known RNA. A merged GTF file is generated based on the transcripts of the samples to be analyzed and the reference GTF file of the corresponding animal [7,108,112]. Following the merged GTF file, only transcripts longer than 200 bp and an ORF transcript length shorter than 300 bp are selected. It also considers the positional relationship in the genome between lncRNA and PCG and designates transcripts consistent with the definition of lncRNA (intergenic, intronic, etc.). Subsequently, it filters only transcripts with low probability using various tools that calculate the potential for transcripts to be coded as proteins. Importantly, tools for evaluating coding potential are diverse and can be used flexibly depending on how to analyze. The transcripts filtered from the sequencing data can be selected and presented as potential novel lncRNA, and can be used for functional analysis and actual lncRNA sequence verification in the future [113–119].
INTERACTION LONG NON-CODING RNA TO mRNA
Among the many known lncRNA functions, a representative and key function is to regulate mRNA expression including , such as epigenetic modification [3,4]. Therefore, a method of conducting mRNA and lncRNA analysis is actively used as an experimental design for exploring animal traits. Importantly, it is used in a wide variety of fields, including production traits [120–123] and quality traits [124–127], milk production [128–130], egg production [9,131,132], stress [133–135], diseases [136] and reproductive traits [137–140].
The types of lncRNA that regulate transcription can be divided into two based on the transcription site and functional location of the lncRNA. It is classified as cis-acting lncRNA if its functional location depends on the transcription site, and trans-acting lncRNA if transcribed to exert functions elsewhere without relying on the transcription site (Fig. 3) [141]. Notably, a method for obtaining a candidate target gene for cis- and trans-acting lncRNA has not yet been fully established. However, the candidate target gene interacting with cis-acting lncRNA is primarily a candidate group of PCGs within 100 kb on the same chromosome of lncRNA. In contrast, the candidate gene interacting with trans-acting lncRNA is a candidate group of PCGs on different chromosomes [141,142].
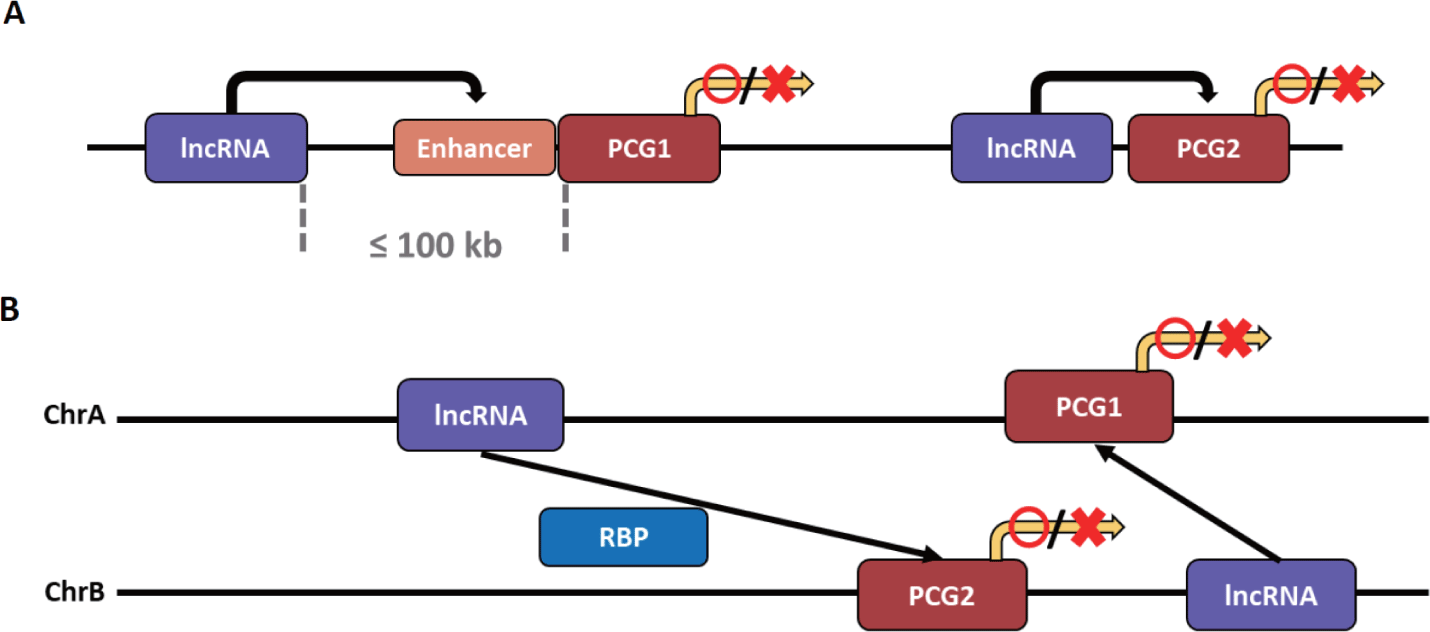
As mentioned earlier, the cis-acting mechanism is preferred because lncRNA is less likely to function normally due to dilution from diffusion and transport to other cellular compartments. After all, the expression level is generally relatively low [141].
The cis-acting lncRNA can increase or inhibit the expression of target genes through various mechanisms. The mechanism by which cis-acting lncRNA increases gene expression is closely related to enhancers. These lncRNAs can be broadly divided into two categories: 1) lncRNAs derived from and transcribed from the enhancer after mutation or translocation has occurred in the gene enhancer [143,144], and 2) those transcribed from other sources that act like the enhancer of the target gene or affect the enhancer [145,146]. These both lncRNAs can activate the target gene by influencing the target gene’s enhancer or act as the enhancer and activate the target gene. As a first mechanism, lncRNA transcripts regulate enhancer activity by forming or maintaining chromatin loops with target genes [147,148]. Additionally, since the lncRNA transcript affects the nuclear localization of the enhancer, it can increase the expression of the target gene by giving strength to the enhancer as an indirect mechanism [149]. The cis-acting lncRNA can activate the expression of a target gene by influencing the enhancer through mechanisms other than spatial interaction. It is an lncRNA that attracts a protein that enhances the enhancer of the target gene [150–152]. There are also cis-acting lncRNAs that activate gene expression independent of enhancers. The lncRNA is transcribed near the target gene, or the preformed chromatin loop structure locates the lncRNA near the target gene, thereby increasing the expression of the target gene by attracting activating factors to the lncRNA [153].
The cis-acting lncRNA not only increases the expression of a target gene but also inhibits it. First, lncRNA near the target gene can suppress the expression by silencing the target gene’s promoter through the enhancer competition of the target gene [154,155]. In addition, the lncRNA is transcribed near the target gene, or the preformed chromatin loop structure places the lncRNA near the target gene so that the lncRNA attracts repressive complexes such as Polycomb repressive complex 2, resulting in the same effect as histone modification. Thus, gene expression can be inhibited [156]. Another mechanism by which cis-acting lncRNA suppresses gene expression is transcriptional interference. Through nucleosome remodeling, in which nucleosomes are rearranged, nucleosome occupancy is reduced, or multiple epigenetic modifications, lncRNA that approaches or overlaps the target gene suppresses the expression of the target gene [157,158].
Previous studies have revealed that cis-acting lncRNA does not only interact one-to-one with the target gene. One lncRNA may be involved in the transcription of several target genes, and conversely, it appears that several lncRNAs may be involved in transcribing a target gene in unison. [4,141].
Unlike cis-acting lncRNAs, trans-acting lncRNAs can interact independently of complementary sequences for target gene regions [99]. Trans-acting lncRNAs function by binding to proteins, DNA, and other RNAs [159]. First, trans-acting lncRNAs can act as post-transcriptional regulatory factors by interacting with RNA-binding proteins (RBPs). These lncRNAs interact with RBPs to inhibit mRNA splicing and the stability and translation of mRNAs [160–162]. Notably, splicing regulation by lncRNA causes a mutation or transformation in the splicing regulation sequence of the target pre-mRNA, resulting in the mis-splicing of the mRNA [163].
Trans-acting lncRNAs can also promote or inhibit the stability of mRNA by interacting directly with RNA through base pairing. This is likely due to the ability to attract proteins involved in mRNA decomposition by directly base pairing with other RNAs [164,165]. While its existence has been revealed and its importance as a post-transcriptional control factor has emerged, research on trans-acting lncRNA is insufficient. Further research will be needed to clarify the apparent correlation between trans-acting lncRNA and target genes and reveal the mechanisms by which several trans-acting lncRNAs interact with RBP.
ANOTHER FUNCTION OF LONG NON-CODING RNA
Until recently, interactions between ncRNAs have rarely been studied. However, recent studies have shown that lncRNA can interact with miRNA and mRNA [166]. Importantly, the lncRNA acts as a sponge to attract miRNA and competes with PCG, which was supposed to bind to miRNA. This attraction process reduces the target gene regulation effect of miRNA [167–170]. Therefore, studies on high-dimensional access to specific biological information are being conducted by analyzing the correlation and interaction of 3 RNAs of lncRNA-miRNA-mRNA [171–173]. It has also been suggested that some lncRNAs can be preferentially post-processed into snoRNA [99,174,175]. As mentioned earlier, the possibility of interaction between lncRNAs or other RNAs is still open, such as various lncRNAs involved in regulating one mRNA expression. However, further research is needed as it is unclear. If these mechanisms are revealed, not only will we be able to understand the principles of lncRNA and mRNA interaction that have not yet been accurately identified, but we will also be able to make much more expansive use of MOI network research using lncRNA.
CONCLUSION
In the past, only studies on mRNA encoded by functional genes were conducted, but now the role of ncRNAs has been re-examined, and research on this topic is being actively conducted. Among them, lncRNA has a high probability of being present in many different places on the genome, and it is known that it has many functions. Therefore, its importance is emerging from these added investigations. As a key function of lncRNA, it can regulate gene expression through various mechanisms. In addition, since it has tissue-specific and species-specific characteristics, it is possible to analyze bioinformation using lncRNA from multiple perspectives in particular tissues of different species. This means that lncRNAs can be used as biomarkers involved in improving reproductive traits and diseases in mammals, including livestock animals. Therefore, lncRNA exploration and functional analysis are being conducted to study various animal traits. However, analysis for identifying lncRNA in animal species other than humans and mice is still lacking, and analysis of the mechanism and function of lncRNA is insufficient. If studies that can supplement these areas are conducted, it is likely that high-dimensional MOI analysis using lncRNA will be possible.