INTRODUCTION
Alongside pork and beef, poultry meat is a major source of protein. According to the Organization for Economic Cooperation Development (OECD) and Food and Agricultural Organization (FAO), poultry meat is expected to account for 41% of the world’s meat supply by 2030 [1]. This reflects the lower price of poultry compared to other meats and increasing consumer preference for white meat, which is recognized as a healthy food item.
In Korea, the demand for chicken is increasing due to the trend toward the consumption of high-quality food as part of a healthy lifestyle [2]. Especially, Korean native chicken (KNC) has a different texture and taste compared to commercial broilers and layers and is highly favored by consumers. KNC has a higher amount of flavor-related components such as inosine, glycine, alanine, and proline than broiler [3]. In addition, KNCs are known to be superior to commercial chickens in physicochemical properties such as water holding capacity, tenderness, and fatty acid composition. [4,5]. For these reasons, consumers are more likely to consume KNCs even though they are more expensive. Therefore, there is a need to develop poultry products satisfying the quality and functionality requirements of consumers.
The quality and flavor of meat are determined by physiochemical properties as color, water holding capacity, tenderness, and other sensory evaluation. These traits were also affected by a complex array of substances such as free amino acids, nucleic acids, lipids, and minerals [6]. Fatty acids are among the most important contributors to meat flavor. Numerous flavor characteristics arise from volatile fatty acid degradation by lipid oxidation during the cooking process [7,8]. Lipid-derived volatiles react with other flavor compounds, such as the products of thermal oxidation and Maillard reaction, to form flavors.
Several studies have been published on candidate genes related to fatty acid composition, mainly in pigs and cattle [9-12]. The major candidate genes related to fatty acids are FASN, SCD, FABP2, and ELOVL7. It has reported that these genes were involved in the synthesis, elongation, and transportation of fatty acids. On the other hand, there have been few studies of the genetic association between fatty acid composition and meat quality in chickens [13]. According to Munyaneza et al. [14], no genome-wide association study (GWAS) has been reported on fatty acid composition in chickens although it is an important determinant of healthy meat.
Therefore, the aim of this study was to determine the candidate genes associated with fatty acid composition in chickens using a crossbred population (between two breeds with opposite phenotypes).
MATERIALS AND METHODS
This research was approved by the Institutional Animal Care and Use Committee (IACUC) of Chungnam National University (202103A-CNU-061). All experiments were conducted following relevant guidelines and regulations.
A total of 767 birds in an F2 crossbred population between Yeonsan Ogye (YO) and White Leghorn (WL) were used for this study. The YO, which is one of KNC breed, is characterized by black feathers, skin, and bones, and has a unique meat flavor, whereas WL is a well-known layer breed with the opposite phenotype of the YO. The F2 generation was created by a reciprocal cross; one WL male was mated with five YO females (B line) and one YO male was mated with five WL females (L line). Using the F2 population for genetic association studies via reciprocal crossbreeding across breeds with opposing phenotypes has the advantage of using normalized data by increasing the variance of the phenotype. This allows for a more accurate estimation of the effect of the genetic variation associated with the phenotype.
All birds were raised on farms in the Animal Genetic Resources Research Center, National Institute of Animal Science (NIAS, Korea) under the same environmental conditions.
The breast meat of birds slaughtered at 10 weeks of age was analyzed. The chicken carcass samples were rapidly frozen at −35°C after slaughter and then stored at −20°C. After 2–6 weeks, samples were transported for experiments and stored at −80°C until deboning. Then, the breast meat was separated from carcass samples, which were thawed at 4°C for 20 h. The lipid was extracted from the breast meat, and it was sequentially mixed with pyrogallol solution, triundecanoin as an internal standard, and hydrogen chloride solution. Next, diethyl ether and petroleum ether were added, respectively, and the weight difference of the total amount was checked to calculate the content of crude fat. Following methylation, the samples were subjected to gas chromatography examination on an Agilent 6890 Gas Chromatograph (Santa Clara, USA). The concentration of each fatty acid was calculated with the internal standard, and each measured fatty acid value was divided by the total fatty acid value and expressed as a percentage.
A total of 30 fatty acids (%) were classified as follows: total saturated fatty acids (SFA), total monounsaturated fatty acids (MUFA), total polyunsaturated fatty acids (PUFA), total omega-3 (ω-3) and total omega-6 (ω-6). The calculations for the various fatty acid groups were as follows: SFA = C10:0 + C12:0 + C14:0 + C16:0 + C18:0 + C20:0 + C22:0 + C24:0; MUFA = C14:1 + C16:1 + C17:1 + C18:1t + C18:1n-7 + C18:1n-9 + C20:1 + C22:1n-9 + C24:1n-9; PUFA = C18:2t + C18:2n-6 + C18:3t + C18:3n-3 + C18:3n-6 + C20:2 + C20:3n-3 + C20:3n-6 + C20:4n-6 + C20:5n-3 + C22:2n-6 + C22:5n-3 + C22:6n-3; ω-3 = C18:3n-3 + C20:3n-3 + C20:5n-3 + C22:5n-3 + C22:6n-3; and ω-6 = C18:2n-6 + C18:3n-6 + C20:3n-6 + C20:4n-6 + C22:2n-6. P/S was the ratio between PUFA and SFA, and ω-6/ω-3 was the ratio between ω-6 and ω-3.
Before conducting a GWAS, all phenotype data were processed to remove skewness and ensure a normal distribution. Data normality was analyzed using the Shapiro-Wilk test, and the following transformation methods were applied: log, square root, sign, square, and cube. The data with the highest p-values were included in the analysis.
Genomic DNA was extracted from blood samples of birds at 8 weeks of age using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). The DNA samples were genotyped using the Illumina chicken 60K BeadChip. PLINK 1.9 software [15] was used for quality control (QC) based on three cut-offs: genotyping rate < 90%, minor allele frequency < 1%, and Hardy-Weinberg equilibrium (HWE) at p < 0.000001. After QC, 29,175 single nucleotide polymorphism (SNP) markers were subjected to the GWAS. Furthermore, principal component analysis (PCA) was conducted to confirm genetic relatedness and the potential for population stratification prior to the GWAS.
The GWAS was conducted on all genotyped SNPs and fatty acids using a mixed linear model (MLM). The MLM was developed with sex (male or female), line (B or L), body weight (8 weeks of age), and the top two principal components as covariates. All analyses were performed using the MLM leaving-one-chromosome-out (MLMA-LOCO) analysis option of the Genome-wide Complex Trait Analysis (GCTA) software package [16]. The model equation was as follows:
where y is the phenotype for fatty acids; X and Z are incidence matrices for parameters b and μ, respectively; b is the vector of fixed effects, including covariates; μ is the vector of SNP effects; g̅ is the accumulated effect of all SNPs except those on the chromosome where the candidate SNP is located; and e is the vector of the residual effect.
Variance components were estimated using the restricted maximum likelihood (REML) option in GCTA for calculating genomic heritability.
After GWAS, significant SNPs were determined based on the Bonferroni-corrected p-value (α = 0.05). We searched for candidate genes in 1 Mb regions around SNPs that were significant in the GWAS and could be involved in the observed significant associations with the phenotypes. The gene annotation process was performed by searching the Ensembl (https://asia.ensembl.org) and National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) databases based on the chicken reference genome (GRCg6a).
RESULTS
A statistical summary of the fatty acid composition in F2 crossbred chickens is provided in Table 1. C18:1n-9 (21.96%) showed the highest content, followed by C16:0 (19.85%) and C20:4n-6 (17.87%). This concurred with reports of oleic acid (C18:1n-9), palmitic acid (C16:0), and linoleic acid (C18:2n-6) being the principal fatty acids in KNCs [3]. In addition, the arachidonic acid (C20:4n-6) content was high, which accorded with the report of flavor-related components being higher in KNCs than general broilers by Jin et al. [17].
The GWAS identified significant SNPs in 15 traits: C10:0, C14:0, C18:0, C18:1n-7, C18:1n-9, C18:2n-6, C20:0, C20:2, C20:3n-6, C20:4n-6, C20:5n-3, C24:0, C24:1n-9, MUFA and PUFA. As shown in Fig. 1, the significant SNPs were mainly located at GGA 10.
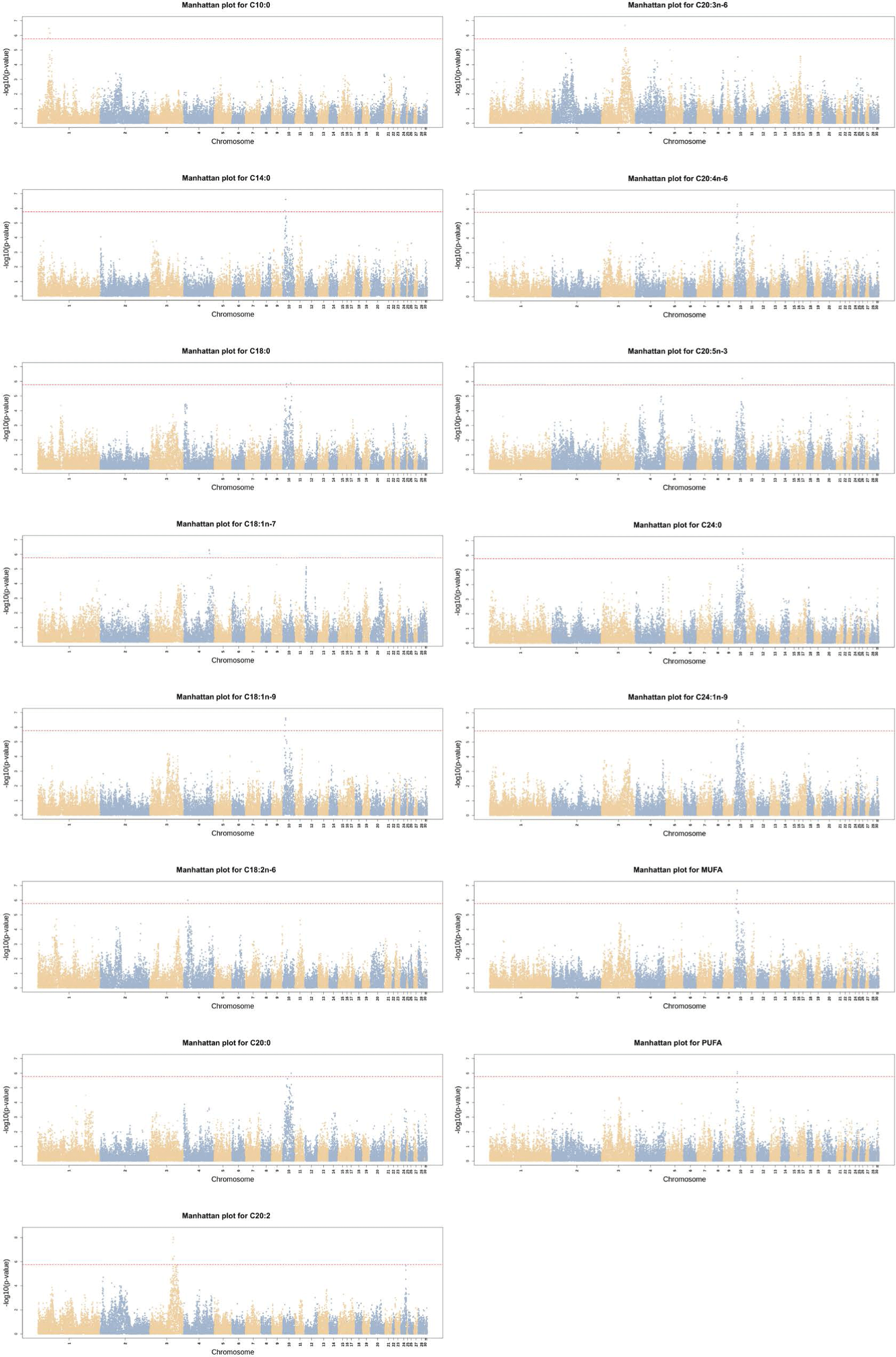
Table 2 lists candidate genes based on the top two SNPs identified for each trait. The same SNP (GGaluGA067637) was observed in C14:0, C18:1n-9, C20:4n-6, and PUFA, and the gene located in this SNP region was also confirmed in MUFA. Gga_rs14381780 in C20:2 and C20:3n-6, GGaluGA070911 in C20:0 and C24:0, and Gga_rs15572763 in C18:0 and C24:1n-9 were also identified.
The candidate genes in C14:0, C18:1n-9, C20:4n-6, PUFA, and MUFA were MCEE and TRPM1, and those in C20:2 and C20:3n-6 were ELOVL4, CYB5R4, and ME1. We also identified the ACSS3 and BTG1 genes in C10:0, the PPARGC1A gene in C18:1n-7, and the ACSL4 gene in C18:2n-6.
Heritability was highest (0.416) in C18:1n-7, but had a low average value (0.169) (Table 3).
DISCUSSION
High arachidonic acid content was observed in YO, a breed of KNC, in this study. A previous study reported that the arachidonic acid content of KNC breeds was significantly higher than that of broilers [18].
Chicken muscle contains more PUFAs than lamb and beef [19]. Hence, more unsaturated volatile aldehydes are produced in chicken compared to other species. These compounds are known to affect the flavor of the chicken. In chicken, the main unsaturated fatty acids are oleic acid and linoleic acid, while arachidonic acid is abundant in phospholipids. Arachidonic acid affects the flavor of meat by forming aromatic compounds such as trans-4, 5-epoxy-(E)-2-decenal, 1-octen-3-one, (E,Z)-2,4-decadienal, (E,E,Z)-2,4,7-tridecatrienal, and hexanal through oxidation [20].
In this study, the potential to explain the fatty acid content of YO using a crossbred group with WL (to enhance the GWAS) was limited. Nevertheless, considering the lack of previous reports on the fatty acid content of YO, the results should provide useful guidance for further research on the unique flavor and meat quality of YO.
In this study, the content of oleic acid, which is one of the main fatty acids in chickens, was high. Moreover, low arachidonic acid content was observed in individuals with a high oleic acid content. A negative correlation between the two fatty acids has also been reported in humans and mice [21,22].
The mechanism underlying the relationship between the two fatty acids has not yet been elucidated. One possible mechanism involves changes in the relative content of other relevant fatty acids. According to Høstmark and Haug [23], α-linolenic acid content is positively related to the ratio of oleic acid to arachidonic acid. In our GWAS, the candidate gene associated with linoleic acid was ACSL4, which causes arachidonic acid catalysis [24].
Another possibility is that oleic acid could act as an inhibitor of Elongase-5 or Delta-5/6 desaturates, which synthesize arachidonic acid. Alternatively, arachidonic acid may act as an inhibitor of Delta-9 desaturate, which produces oleic acid. Further research is needed to validate this hypothesis, although it is supported by previous studies showing that PUFAs of the ω-3 group, such as α-linolenic acid, inhibit this transcription [25].
The MCEE gene, which was common in C14:0, PUFA, and MUFA, encodes a methylmalonyl-CoA epimerase and is involved in fatty acid catabolism [26]. In addition, the TRPM1 gene forms a protein constituting the transient receptor potential (TRP) cation channel, which plays an important role in fatty acid oxidation and signal transduction pathways [27,28]. According to Khan et al. [29], the amount of volatile compounds derived from fatty acid metabolism is increased by products formed as a result of enzymatic activity occurring during the metabolic process. Therefore, the two candidate genes identified by our GWAS may have contributed to the metabolic process of each fatty acid, and influenced the production of volatile compounds that determine the meat quality and flavor of chicken.
Candidate genes that were common in C20:2 and C20:3n-6 included ELOVL4, CYB5R4, and ME1. The ELOVL4 gene encodes an enzyme required to synthesize long-chain fatty acids and plays an important role in the formation of long-chain PUFAs [30]. According to Duckett and Kuber [31], oxidation products formed from long-chain unsaturated fatty acids, such as ω-3 and 6, have a significant effect on the flavor of lamb. CYB5R4 is an electron donor for fatty acid desaturase by stearoyl-CoA desaturase (SCD) [32]. SCD plays a key role in the formation of double bonds and contributes to the biosynthesis of unsaturated fatty acids, such as palmitoleic acid (C16:1) and oleic acid (C18:1n-9), from SFAs such as palmitic acid (C16:0) and stearic acid (C18:0). According to a study by Kawaguchi et al. [33], it was confirmed that the CYB5R4 gene was present in the quantitative trait loci (QTL) region associated with the oleic acid percentage in Japanese Black cattle. In addition, ME1 encodes an enzyme that generates nicotinamide adenine dinucleotide phosphate (NADPH) for fatty acid biosynthesis, and it has been reported that overexpression of endogenous ME1 promotes SFA and PUFA biosynthesis [34].
Candidate genes identified in C10:0 included ACSS3 and BTG1. ACSS3 encodes acyl-CoA synthetase and activates short-chain fatty acids. According to Dinh et al. [8], short-chain fatty acids, which are major volatile substances produced at high temperatures along with lactones, alcohols, and ketones, are among the important factors determining the flavor of meat. Because they react with other compounds during cooking, including participation in the Maillard reaction, they can produce more desirable volatiles compared to autoxidation. Also, according to Buitenhuis et al. [35], the ACSS3 gene was identified as a candidate gene affecting the content of C10:0 in cattle milk fat composition.
The candidate gene identified in C18:1n-7 was PPARGC1, which encodes peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) in humans. Nikolić et al. [36] reported that overexpression of PGC-1α affects the activation of transcription factors that convert muscle cells into oxidative metabolism and increases the mRNA expression of genes regulating lipid metabolism.
Currently, the poultry industry in Korea relies mainly on imported foreign species due to their excellent growth ability; such species account for > 90% of the Korean poultry industry. As such, the industry would face industrial collapse if imports became difficult to obtain. Therefore, genome-based analysis of various economic traits of KNCs, as well as of meat-specific components, is urgently needed to discover physiologically functional substances. This could lay a foundation for the development of superior species for meat and enhance the international competitiveness of the Korean poultry industry.
In this study, a GWAS was performed to determine the fatty acids contributing to the unique meat quality and flavor of a breed of KNC. However, the results may not fully explain the relationships of meat quality and flavor with fatty acid composition.
The volatile substances that affect meat quality and flavor are affected by various environmental factors. In general, it is difficult not only to generate flavor defects, but also to improve flavor during manufacturing and processing. The influence of different substances on flavor depends on the cooking method. Cooking for an extended period causes a Maillard reaction, while grilling or frying at ≥ 100°C results in the generation of aromatic components, such as heterocyclic compounds [37]. The nutritional conditions during poultry breeding are also very important factors affecting meat quality and safety. In particular, linoleic acid and α-linolenic acid, which synthesize various fatty acids that affect flavor, are essential fatty acids not produced in the body; therefore, they are greatly affected by intake amounts. In addition, breeding conditions, storage temperature and duration, and the post-slaughter treatment process can cause many changes in meat quality and flavor. Therefore, to explain the genetic relationship between fatty acid composition and meat quality, a more detailed analysis that considers the effects of the above-mentioned environmental factors is required.
Numerous studies of the fatty acid composition and role of individual fatty acids in meat quality and taste have been conducted. However, most of the studies were conducted on ruminants or pigs; few have analyzed poultry meat. In a QTL study of fatty acid content, 1,201 QTL regions in cattle, and 6,460 in pigs, were investigated, whereas in a study on chickens only 10 QTL regions were investigated (Animal QTL Database; based on an accession date in August 2022). Therefore, the results of this study provide important genetic information on the fatty acid composition of poultry meat and help explain their influence on the quality and flavor of poultry meat.
CONCLUSION
A GWAS was performed of the fatty acids responsible for the unique meat quality characteristics of YO, which is a breed of KNC. Significant results were obtained for 15 fatty acids, including oleic acid and arachidonic acid. The functions and mechanisms of candidate genes affecting meat quality and flavor, such as MCEE and TRPM1, were also confirmed. Additional analyses will be required before utilizing polygenic traits, such as fatty acid composition, and can be exploited in actual breeding programs for the development of high-quality breeds. Nevertheless, our study provides genetic information that could lead to the improvement of meat quality, especially the fatty acid composition of KNCs.
















