Lactic acid bacteria (LAB) are known for their ability to convert carbohydrates to lactic acid, which helps to maintain the appropriate environment of the host gut and can have beneficial effects on the health of humans and animals. Lactiplantibacillus plantarum, formerly named Lactobacillus plantarum, can be found in various environments such as dairy products, fermented sausages, meat, fish, vegetables, and human feces. The L. plantarum strain is also present in the gut microbiota of healthy individuals and has been shown to have various health benefits for its host. Some of the benefits include improved digestion, enhanced immune system function, and reduced inflammation [1,2]. L. plantarum L55 is a probiotic strain of bacteria that has been isolated from kimchi and has been found to be highly resistant to acidic conditions and bile salts, making it a good candidate for use as a probiotic supplement and starter culture for yogurt.
The genome of L. plantarum is relatively large, ranging from 2.8 to 3.3 megabases, and exhibits high variability among different strains. The L. plantarum genome encodes for a diverse array of genes, which are involved in a range of biological processes, including carbohydrate metabolism, amino acid biosynthesis, and stress responses. This genetic diversity is thought to contribute to the adaptability of L. plantarum to different environments and its ability to compete with other bacteria.
Using de Man-Rogosa-Sharpe (MRS) medium (Becton, Dickinson, Rungis, France), L. plantarum L55 was grown for 24 h at 37°C. HiGeneTM Genomic DNA Prep kit (BIOFACT, Daejeon, Korea) was used to extract genomic DNA in accordance with the manufacturer’s instructions. We used an Illumina® DNA Prep kit to assemble our short-read sequencing library (Illumina, San Diego, CA, USA). Short-read sequencing (300 base pair [bp], paired-end) was carried out on the Illumina MiSeq platform with the Illumina MiSeq reagent kit V3 (Illumina). The Oxford Nanopore Ligation Sequencing kit was used for the library preparation of long-read sequencing (Oxford Nanopore, Oxford, UK). The MinION sequencing instrument (Oxford Nanopore) with a MinION flow cell (R9.4.1) was used for the long-read sequencing (Oxford Nanopore). 675,276 paired-end reads (165,535,022 bp) were obtained from Illumina short-read sequencing, while 251,401 reads averaging 1,342 bases in length were obtained from Oxford Nanopore long-read sequencing.
The de novo genome sequence was assembled using a Flye assembler (v. 2.9) [3] with default options. Adapter sequence removal and quality control of short-reads were performed using TrimGalore (v. 0.6.7) [4] with the “paired” parameter. The quality of the draft genome assembly was improved by correcting errors with Pilon (v. 1.24) [5] with default parameters. Gene predictions for the chromosome and plasmid sequences were performed with Prokka (v. 1.14.5) [6]. An assessment for genome assembly completeness was conducted by BUSCO (v. 5.2.2) [7] using the OrthoDB v10 bacterial (bacteria_odb10) lineage dataset.
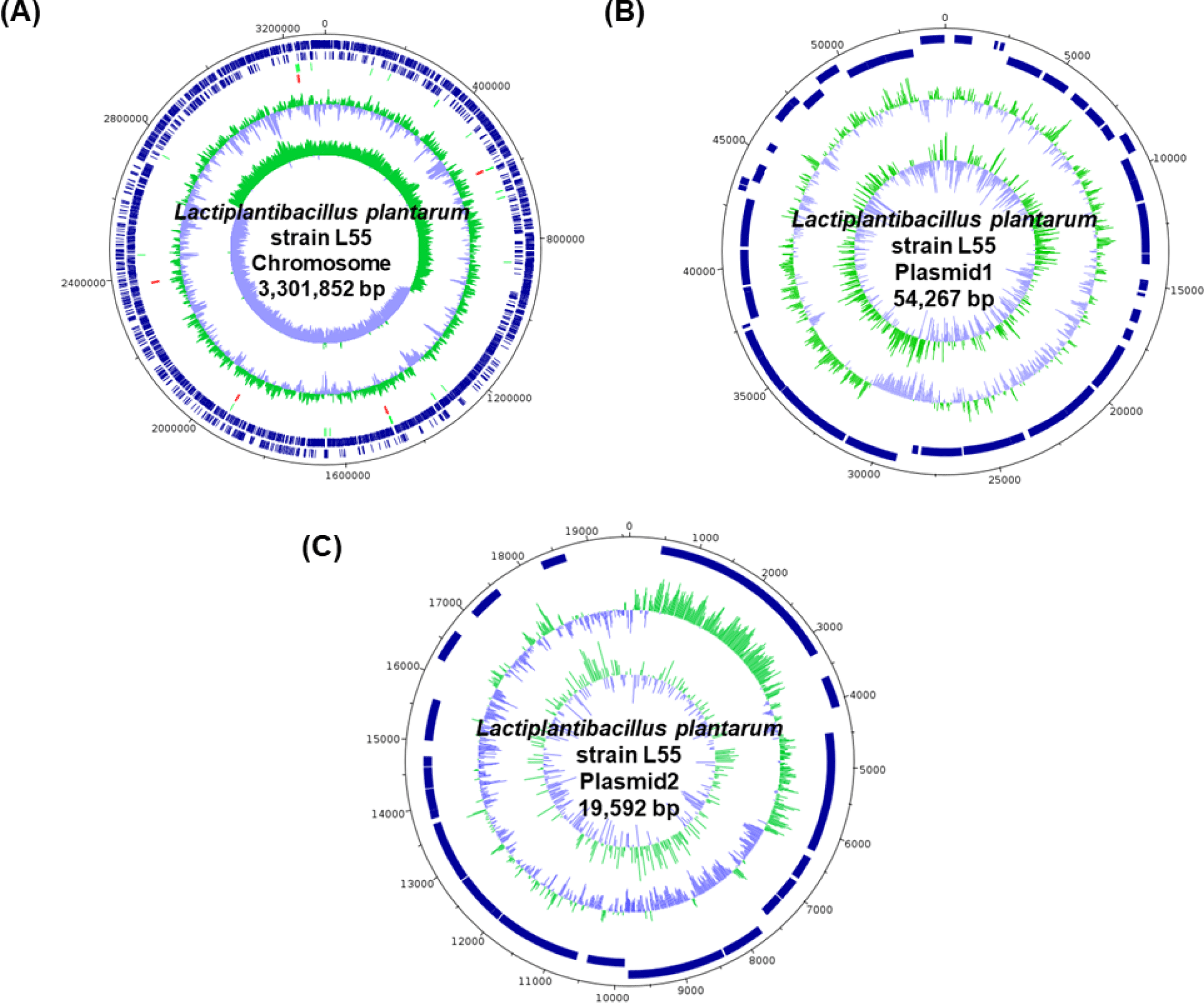
The complete genome sequence of L. plantarum L55 consisted of one circular chromosome (3,301,852 bp) with an overall GC content of 44.5% and two circular plasmid sequences of 54,267 bp and 19,592 bp, respectively (Table 1).
| Length (bp) | GC (%) | Depth | CDSs | tRNA | rRNA | |
|---|---|---|---|---|---|---|
| Chromosome | 3,301,852 | 44.5 | 66.0 | 3,079 | 66 | 16 |
| Plasmid1 | 54,267 | 39.1 | 327.0 | 48 | 0 | 0 |
| Plasmid2 | 19,592 | 40.5 | 405.0 | 22 | 0 | 0 |
| Total | 3,375,711 | 44.4 | 72.2 | 3,149 | 66 | 16 |
The sequences of the genome and the plasmids predicted a total of 3,231 genes, including 3,149 genes that code for proteins, 16 genes for ribosomal RNA, and 66 genes for transfer RNA (Fig. 1).
Functional annotation of the predicted genes was performed with EggNOG-mapper (v. 2.1.9) [8] using EggNOG database version 5.0.2 [9]. Within the total of 3,149 protein-coding genes, biological functions were defined for 2,678 (85.04%). Annotated genes were associated with transcription (290), translation (168), cellular envelope biogenesis (155), energy production (116), secretion (62), signal transduction (53) and secondary metabolite biosynthesis (15). Moreover, each gene was assigned with a putative function.
Folate and riboflavin biosynthesis, molybdopterin biosynthesis, sulfur metabolism, cellulose degradation, terpenoid and polyketide biosynthesis, and bacteriocin production were some of the functions that were postulated to be carried out by L. plantarum L55.
















