INTRODUCTION
Extracellular vesicles (EVs) are nanovesicles that cells secrete into the extracellular environment and are characterized by their bilayer lipid membrane structure. Of these vesicles, the most well-known are exosomes, which are particles typically measuring 30–150 nm [1]. EVs, including exosomes, are found in various body fluids, such as blood, milk, cerebrospinal fluid, and saliva. They play a pivotal role in cell–cell interactions through the nucleic acids, proteins, metabolites, and lipids they contain [2]. These various bioactive substances in the vesicles are transmitted to the recipient cell, and this communication of information affects physiological processes, such as the regulation of immune responses, cell survival or death, and cell development [3]. Because of their potential for physiological regulation, EVs have recently been extensively studied in a range of fields.
One of the main components of EVs are microRNAs (miRNAs), which are small, noncoding RNAs. They are short, about 22 nucleotides (nt) in length, and have been found to play a pivotal role in regulating gene expression, leading to a growing body of research [4]. The complete set of miRNAs in a given cell or biological sample, known as the miRNome, is crucial for understanding the regulatory landscape within EVs [5]. Transported via EVs, miRNAs are involved in intercellular communication and immune regulation [6]. Specifically, miRNAs bind to target mRNAs and lead to their translation inhibition or degradation [7]. A milk-derived miRNA, miR-22-3p, has been reported to promote proliferation of human-derived intestinal epithelial cells [8]. Moreover, miR-31-5p enhanced endothelial cell proliferation and angiogenesis as well as alleviated diabetic wounds [9]. Understanding the miRNA profiles in EVs can provide insights into their roles in disease mechanisms, immune responses, and developmental processes.
Previous studies have reported that milk-derived EVs could affect intestinal epithelial cells. Exposure of milk EVs to HT-29 cells confirmed the uptake of milk EVs by these cells, accompanied by an elevation in miRNA levels within the EVs [10]. Exosomes derived from mouse breast milk promoted viability and proliferation of mouse intestinal epithelial cells [11]. Furthermore, EVs derived from human milk were reported to promote ZO-1 expression in Caco-2 cells and suppress the expression of inflammation-related genes [12]. These findings highlight the therapeutic potential of milk-derived EVs to promote gut health and modulate immune responses as well as the critical need for further comparative studies on EVs from different breeds or lactation stages to fully understand their biological functions and applications.
Colostrum, the milk secreted shortly after delivery, is important for the immunity and development of newborn animals, and its properties have been shown to be distinct from those of mature milk, and so are the properties of EVs. A comparison of EVs from colostrum, first milk, and mature milk showed that colostrum and first milk have more EVs than mature milk [13]. Differentially expressed milk-derived EVs across different phases and revealed the differences between human milk and bovine milk [14]. Colostrum- and mature milk–derived EVs have been shown to be able to protect intestinal epithelial cells from lipopolysaccharide (LPS)-induced damage and inflammation. Specifically, EVs from colostrum and mature milk have been found to exert differential effects on the expression of apoptosis-related and proinflammatory genes as well as to enhance cell proliferation and intestinal barrier function [15].
Holstein is the main breed for milk consumption, whereas Jersey is a recently introduced breed in Korea and is known for producing high-quality dairy products [16]. Although there have been several studies comparing the composition of milk between the two breeds, no comparative analysis of the characteristics of milk-derived EVs and miRNAs has been conducted. Therefore, this study aimed to compare the miRNAs and characteristics of colostrum- and mature milk–derived EVs from Holstein and Jersey breeds and to determine how EVs affect intestinal epithelial cells via transcriptomic analysis.
MATERIALS AND METHODS
All colostrum and mature milk samples were collected from three Holstein and three Jersey cows within 3 days and 1 month after delivery, respectively. To ensure comparability and control for breed-specific differences, these cows were raised on the same facility and fed the same diet. The pooled samples from each breed were immediately frozen and stored at −80°C until further analysis.
EVs were isolated from each milk sample using the previously described method, with slight modifications [17]. Briefly, the milk samples were centrifuged at 1,500×g for 10 min to remove the fat. Subsequently, the supernatant was centrifuged at 16,000×g for 1 h to pellet the cell debris. To obtain the whey fraction, the supernatant was centrifuged for an additional 1 h at 50,000×g. The whey fraction was ultracentrifuged at 100,000×g, and then the resulting supernatant was centrifuged at 135,000×g for 90 min to pellet the EVs. The centrifugation step was repeated, and phosphate buffered saline (PBS) was used to wash the EVs to ensure purity. The EVs were resuspended in PBS and stored at −80°C until further analysis.
From each EVs, miRNAs were extracted as indicated using a QIAzol reagent (Qiagen, Hilden, Germany) and the miRNeasy Mini Kit (Qiagen). The purity and integrity of the extracted miRNAs were measured using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Subsequent library generation and sequencing were performed at Macrogen (Seoul, Korea). The SMARTer smRNA-Seq Kit for Illumina was used for library construction and sequenced using the HiSeq 2500 System (Illumina, San Diego, CA, USA). The generated raw data were filtered based on quality. The reads were sequentially aligned to reference genome, miRBase version 21 [18] and noncoding RNA database, RNAcentral 10.0. This alignment allowed for the classification of known miRNAs as well as other types of RNA, including tRNA, snRNA, and snoRNA. Novel miRNA prediction was performed using miRDeep2 [19]. The read counts for each miRNA were extracted from mapped miRNAs to report the abundance of each miRNA. Differentially expressed miRNAs (DEM) were determined by comparing across conditions each miRNA using edgeR of the R package with thresholds of |log2 fold change > 1| and p-value < 0.05. Representative miRNA target–binding sites were predicted using the TargetScan database (version 7.0). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations were analyzed using the DAVID online tool (https://david.ncifcrf.gov/).
HT-29 cells obtained from ATCC were grown in RPMI 1640 medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum. Complete monolayers of HT-29 cells were washed three times with PBS. Then, Holstein colostrum–derived EVs (HC;100 μg/mL) were added to the medium and incubated for 6 h at 37°C in the presence of 5% CO2. After exposure, the cells were washed with PBS, adherent cells were detached using a cell scraper, and total RNA was extracted using the RNeasy Mini Kit (Qiagen). The quality of total RNA was measured using the Agilent 2100 Bioanalyzer. The extracted RNA was stored at −80°C until further analysis.
For RNA-seq, libraries were prepared using the TruSeq RNA Sample Prep Kit version 2 (Illumina) according to the manufacturer’s protocol, and paired-end sequencing was performed using the Illumina HiSeq 2000 instrument. Sequenced raw data were trimmed using Trimmomatic 0.38 to remove adapter sequences, bases with quality < 3 from the ends of the reads, and bases that did not meet window size = 4 and mean quality = 15 using the sliding window trim technique. The trimmed data were generated by removing reads shorter than min length = 36 bp, and subsequent analysis were conducted using high-quality reads that based on the established quality controls. The Hisat2 version 2.1.0 program was used to build an index of the reference genome and paired-end clean reads of the human genome reference (GRCh38) were read and compared. Uniquely mapped reads were quantified with Subread/featureCounts version 1.5.1 using ENSEMBL version 82 transcriptome definitions. The generated data were subjected to differential expression analysis between samples using the R package edgeR, with genes corresponding to the thresholds |log2 fold change > 1| and p-value < 0.05 defined as significantly differentially expressed genes (DEG). GO and KEGG pathway annotations were analyzed using the DAVID online tool.
The identified miRNA profiles were visualized using EVenn (http://www.ehbio.com/test/venn). Statistical analyses and visualization were performed using GraphPad Prism 9.0 (San Diego, CA, USA).
RESULTS
To determine the miRNA profile of EVs present in each raw milk sample, EVs were isolated from each sample via ultracentrifugation. The miRNAs were extracted from the isolated EVs, and small RNA sequencing was performed using the HiSeq platform of Illumina (Fig. 1). A total of 421 miRNAs were identified in HC, and 259 miRNAs were sequenced in Holstein mature milk–derived EVs (HM). A total of 233 miRNAs were common to HC and HM, with 188 miRNAs identified in colostrum only and 26 miRNAs identified in mature milk only. Meanwhile, 198 and 282 miRNAs were identified in Jersey colostrum–derived EVs (JC) and Jersey mature milk–derived EVs (JM), respectively. There were 182 miRNAs that were common to both Jersey colostrum and mature milk, with 16 and 100 miRNAs identified in JC and JM, respectively.
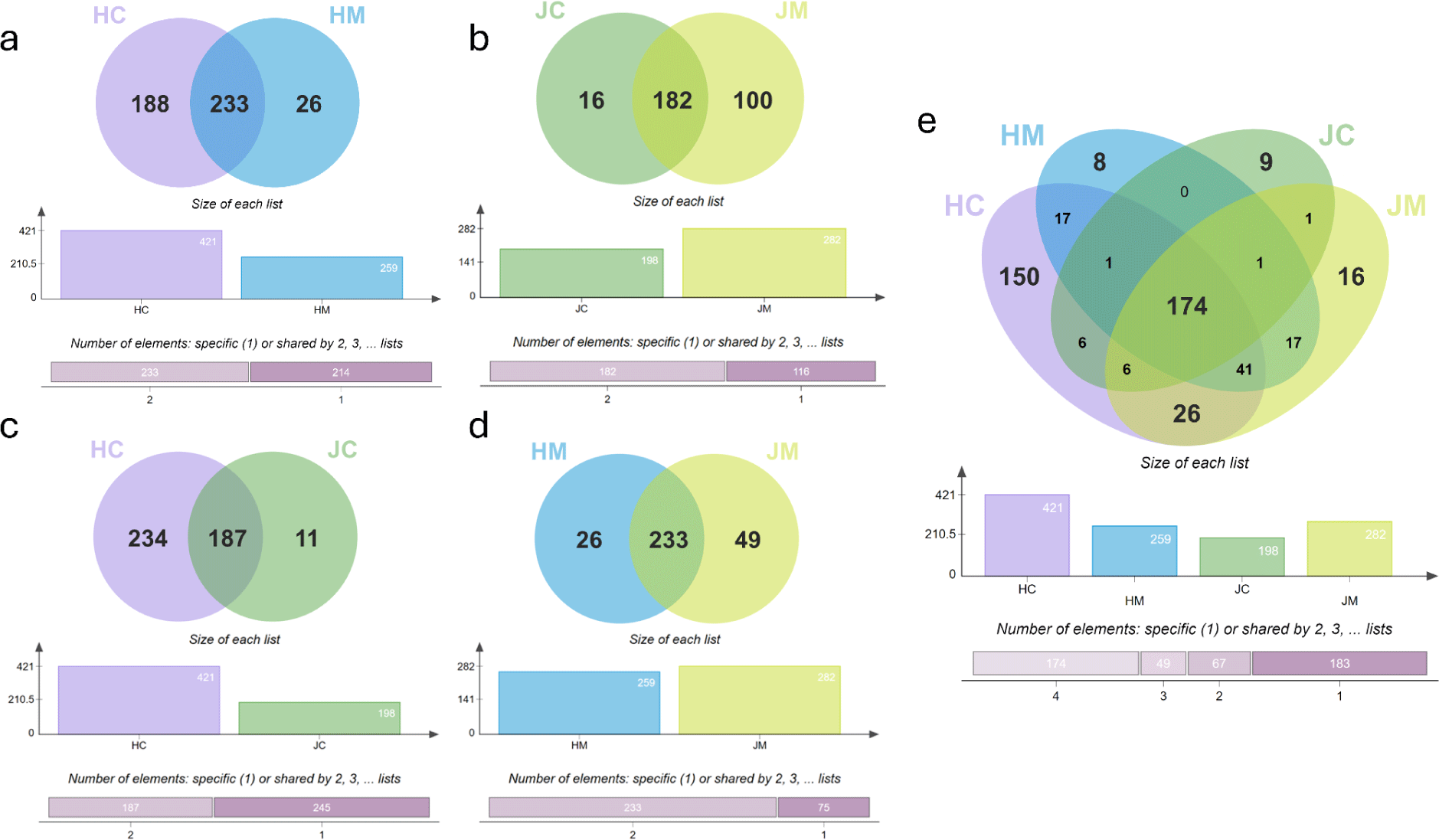
Next, the differences in miRNA profiles between the species were compared. The comparison revealed that 187 miRNAs were commonly expressed in both samples whereas 234 and 11 miRNAs were unique to HC and JC, respectively. Similarly, comparison between HM and JM revealed 233 shared miRNAs, with exclusive counts of 26 in HM and 49 in JM. Finally, comparison of the miRNA profiles of all samples revealed that 174 miRNAs were common across all samples. Unique miRNAs detected numbered 8, 9, and 16 in HM, JC, and JM, respectively. Contrarily, 150 miRNAs were exclusively found in HC, indicating that Holstein colostrum possesses the most distinct and extensive miRNA profile among the samples.
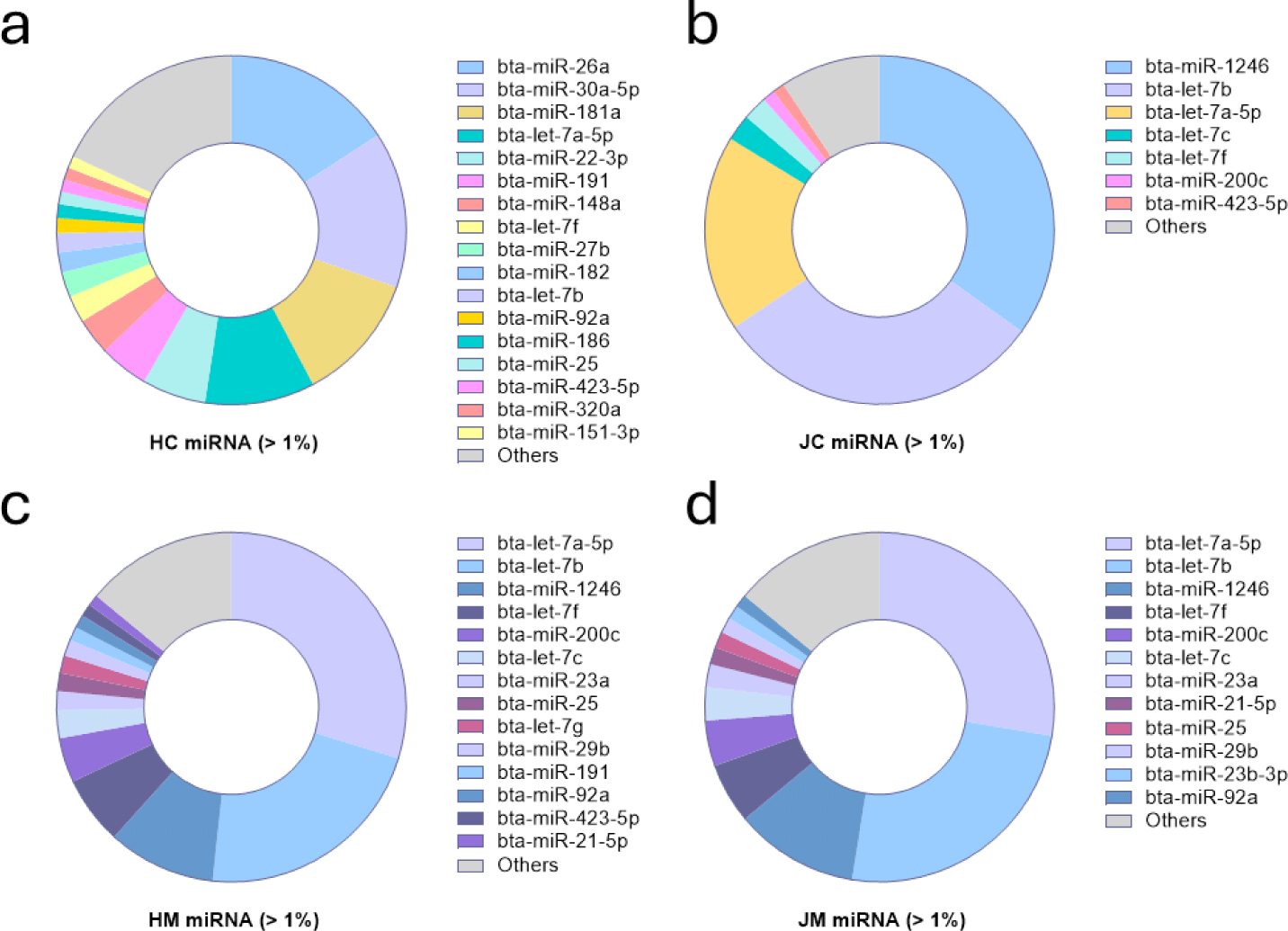
Identification of miRNAs with expression levels exceeding 1% in each sample showed that 17 miRNAs were evenly expressed across the HC samples, with the top 7 accounting for about 66% of the total (Fig. 2). Meanwhile, seven miRNAs in JC had expression levels greater than 1%, whereas the two most highly expressed miRNAs accounted for about 65%. In mature milk EVs, 14 miRNAs in HM and 12 miRNAs in JM had expression levels greater than 1%. Interestingly, the top 6 miRNAs were the same across all samples except HC, although their proportions differed. In addition, the top 7 miRNAs in HC and JC were identical, comprising 76% and 78% of the total miRNA expression, respectively.
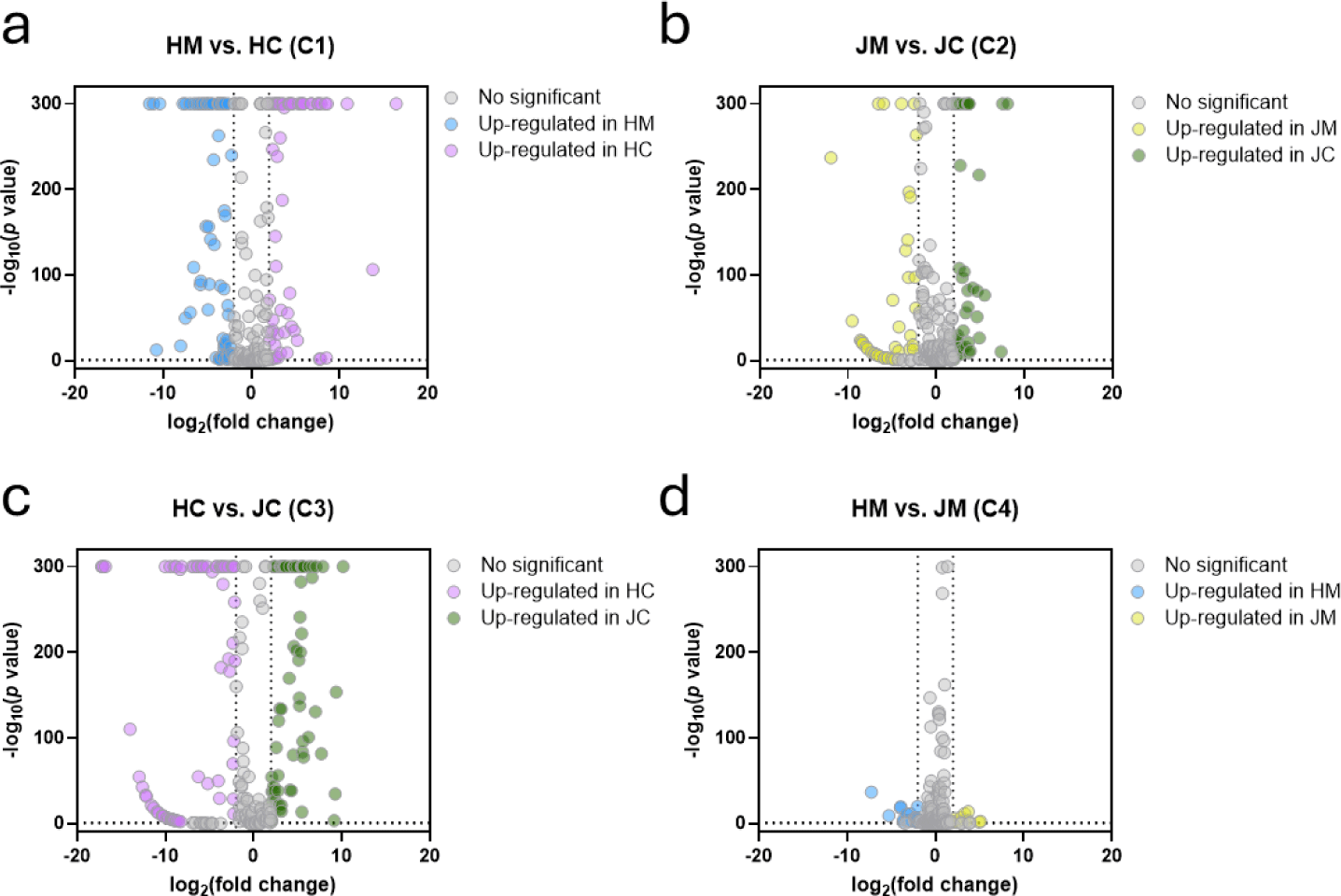
To further analyze the miRNA profiles among samples, the identified miRNAs were filtered and normalized. As a result, 570 of 793 mature miRNAs were excluded, and 223 mature miRNAs were used for statistical analysis (Fig. 3). In comparison group C1, 60 miRNAs were upregulated and 67 were downregulated in HC compared with HM. In comparison group C2, 39 miRNAs were upregulated and 48 were downregulated in JC compared with JM. Interestingly, 68 miRNAs were upregulated and 70 were downregulated in JC compared with HC (C3). Contrarily, the comparison between HM and JM showed 9 upregulated and 20 downregulated miRNAs (C4). This suggests that the DEM between colostrum and mature milk are more pronounced in Holstein cows than in Jersey cows and that the species-specific differences are more evident in colostrum compared with mature milk.
Gene enrichment analysis was conducted to predict the biological activities associated with the DEM for the three remaining combinations, except for C4, the mature milk comparison, which exhibited a slight difference in expression. The target genes of the top 10 DEM in each sample were predicted by TargetScan, and enrichment analysis of the selected gene set was conducted using the DAVID tool (Table 1). Immune-related pathways were found to be regulated in most comparative combinations. However, in both Holstein and Jersey breeds, these pathways were more prominently regulated in colostrum than in mature milk. In addition, comparison between JC and HC showed that more immune-related pathways were regulated in HC. Moreover, KEGG pathway analysis revealed that pathways related to MAPK signaling pathway and FoxO signaling pathway were upregulated, and cancer-related pathways were found to be upregulated in C1 and C2 rather than in C3. The top 10 pathways associated with immune responses are shown in Tables 2, 3, 4, 5, 6, and 7.
DEMs, differentially expressed miRNAs; C1, comparison between HC and HM; C2, comparison between JC and JM; C3, comparison between JC and HC; HC, Holstein colostrum–derived extracellular vesicles; HM, Holstein mature milk–derived extracellular vesicles; JC, Jersey colostrum–derived extracellular vesicles; JM, Jersey mature milk–derived extracellular vesicles.
As HC had the most characteristic miRNA profile, GO and KEGG enrichment analyses were conducted on the top 7 miRNAs that accounted for more than 3% of abundance. Among the enrichment results, a total of 37 items related to biological processes, 15 related to cellular components, and 15 related to molecular functions were identified as important. KEGG pathway analysis revealed that a total of 31 pathways were significantly matched, most of which were related to immunity or cancer. The top 10 pathways associated with immune responses are shown in Table 8.
Based on the previous results, HC is the most likely to regulate immune-related functions in the sample. Therefore, to confirm the effect of HC, human colon cell line HT-29 was treated with HC and subjected to transcriptomic analysis. DEG analysis revealed that of a total of 2,892 genes, 203 and 255 were upregulated and downregulated, respectively, upon treatment with HC. Gene enrichment analysis of DEG was conducted, and the results are shown in Table 9. GO analysis revealed that the upregulated genes were related to chemotaxis and negative regulation of the macrophage cytokine production pathway. Downregulated genes were identified as specific genes for negative regulation of platelet aggregation, telomere maintenance in response to DNA damage, regulation of cell cycle G2/M phase transition pathway. KEGG pathway analysis revealed that the upregulated genes did not produce substantial results and that the downregulated genes were associated with the NF-kappa B signaling and metabolic pathways.
DISCUSSION
In this study, the miRNA characteristics of EVs derived from colostrum and mature milk of two cattle breeds were compared. The comparison revealed that the most diverse miRNAs were evenly present in Holstein colostrum EVs whereas fewer miRNAs were identified in Jersey colostrum. For the miRNA profiles of mature milk samples, the top 7 miRNAs were found to be identical in Holstein and Jersey, accounting for approximately 77% of the miRNAs. The DEM analysis revealed more miRNA differences in the comparison between mature milk and colostrum, consistent with the miRNA profile results, and few DEMs were found in the comparison between mature milk miRNAs. Gene enrichment analysis of the target genes of the top 10 DEM revealed that immune-related pathways were involved in colostrum than in mature milk in both Holstein and Jersey breeds. Holstein colostrum EVs with the most distinctive miRNA signatures were further analyzed by transcriptomics for their effects on intestinal epithelial cells, and genes associated with immune-related pathways were found to be modulated. These results indicate that EVs derived from Holstein and Jersey colostrum, rather than mature milk, are enriched in miRNAs that regulate immune-related pathways and that Holstein colostrum is the most likely immune modulator among them.
miRNAs are known to play a pivotal role in biological processes by regulating gene expression. Due to this property, research on the importance of miRNAs is rapidly growing [20–22]. Previous studies have demonstrated that miRNAs in bovine can influence gene expression in humans [10] and are also used to ameliorate diseases or for the diagnosis of diseases as biomarkers [23–25]. The bioactive functions of EVs are closely associated with their internal miRNAs [26]. Therefore, the identification of the profile of miRNAs present in EVs is important for EV characterization.
Most dairy cows currently bred are Holstein, which are characterized by their large body size and higher milk yield compared with other breeds [27]. Jersey cows are characterized by their smaller body size and lower feed consumption compared with Holstein [28]. They are also more heat-resistant than Holstein cows and have a higher amount of protein and fat in their milk, which makes them more efficient in the production of dairy products [16]. Most farmers breed Holstein for efficient milk production, and thus, most of the milk consumed by humans comes from Holstein, but Jersey cows are increasingly being domesticated to produce high-quality dairy products [29]. However, most comparisons of Holstein and Jersey milk have focused on their nutritional composition, and the miRNA content has not been investigated. To the best of our knowledge, this study is the first to explore the profiles of EV-derived miRNAs in colostrum and mature milk between Jersey and Holstein cows. The miRNA profiles revealed that in Holstein breeds, more miRNAs were identified in colostrum-derived EVs than in mature milk EVs. Meanwhile, in Jersey milk, more miRNAs were detected in mature milk–derived EVs than in colostrum-derived ones. In a previous study, miRNA analysis of colostrum and mature milk from Holstein and Doğu Anadolu Kirmizisi cows revealed that in both breeds, known miRNAs were more highly expressed in mature milk than in colostrum [30]. Other studies have investigated miRNAs in bovine and porcine breast milk and detected about 100 more miRNAs in colostrum than in mature milk [31,32].
Jersey milk is known to have higher protein and lipid contents than Holstein milk. Previous studies have demonstrated that milk with higher ratios of fat and protein is enriched in miRNAs that regulate protein and lipid metabolism, mammary gland development, and amino acid biosynthesis [33,34]. However, the present study found fewer differences in miRNAs in the mature milk samples and no significant association with protein and fat metabolism compared with the colostrum samples. A previous study reported that colostrum from Holstein and Jersey breeds did not differ in terms of fat and protein contents [29]. To confirm the association between miRNA analysis results and milk composition, further compositional analysis of milk samples is warranted. Moreover, the results are controversial because the miRNA profiles of colostrum and mature milk have not been sufficiently studied yet, and the expression level of miRNAs may be influenced by the state of the individual from which they are derived, sample management, and time and duration of lactation [35]. To more accurately compare the miRNA profiles, it is important to use a wide range of samples under various conditions.
Previous studies have demonstrated that miRNAs in colostrum are more involved in immunologic pathways than miRNAs in mature milk [36,37]. Analysis of miRNAs in porcine breast milk has revealed that there are more immune-related miRNAs in colostrum and that these miRNAs are present in the serum of piglets [32]. The results of the present study indicated that Holstein colostrum compared with mature milk was associated with biological activities, such as regulation of apoptotic process, negative regulation of cell proliferation, and cell–cell signaling, as well as KEGG pathways, such as MAPK, Ras, and chemokine signaling pathways. Similarly, the target genes of Jersey colostrum miRNAs compared with mature milk were predicted to be associated with biological processes, such as negative regulation of cell proliferation, phosphorylation, negative regulation of Wnt signaling pathway, and KEGG pathways, such as mTOR signaling pathway. Meanwhile, the target pathways of DEMs in HC and JC were predicted to regulate more immune-related processes in HC than in JC. In addition, the KEGG pathway analysis revealed that both upregulated and downregulated miRNAs in each comparison combination would affect different types of cancer and immune-related pathways. These results are consistent with previous studies demonstrating that milk miRNAs are strongly associated with cancer and immunity [22].
The most highly expressed miRNAs in Holstein colostrum include bta-miR-26a, bta-miR-30a-5p, bta-miR-181a, and bta-let-7a-5p. miR-26-a regulates the secretion of proinflammatory cytokines in LPS-stimulated macrophages [38]. Moreover, miR-30a-5p and miR-26a have been reported to be capable of alleviating oncogenic inflammation in the colon by regulating the expressions of NF-κB and IL-6 [39,40]. miR-181a has an anti-inflammatory activity mediated by the inhibition of IL1a in LPS-treated monocytes and macrophages [41]. In addition, let-7a-5p regulates IL-10 secretion in the inflammatory environment; it improved lung function by reducing collagen deposition and macrophage infiltration in mice with acute lung injury [42]. Together with the previous findings, further studies are warranted to confirm the possible anti-inflammatory functions of miRNAs with high proportions in HC and the overall role of HC. Furthermore, miRNAs typically do not bind directly to DNA, but instead inhibit gene expression by attaching to complementary sequences of the target mRNA and inhibiting translation or degrading the mRNA itself. This mode of action carries a low risk of causing direct mutations in DNA sequences [6]. However, the long-term effects of miRNA therapy and whether it may have unintended consequences are still unclear and need further study and long-term observation.
EVs are characterized by their ability to interact with other cells through their internal miRNAs and other bioactive substances. Previous studies have demonstrated that EVs are taken up by target cells by pathways such as phagocytosis, endocytosis, and membrane fusion [43]. Milk-derived EVs have been shown to be taken up by Caco-2 cells in a concentration-dependent manner and are not cytotoxic even after 6 h of EV treatment [44]. It has also been confirmed that milk EVs are taken up by differentiated human macrophages [45]. In this study, the effects of HC on the human colon cancer cell line HT-29 were investigated using a transcriptomics approach. It was found that most of the regulated pathways were associated with immunity, with more downregulation than upregulation. However, there were not many significantly regulated pathways and matched genes, which may be due to the concentration and time of exposure of EVs [10].
CONCLUSION
In conclusion, this study identified differences in the miRNA profiles of HC, HM, JC, and JM, with the latter showing less significant differences between species, whereas more diverse miRNAs were characterized in Holstein than Jersey colostrum EVs. Interestingly, regardless of the species, more immune-related pathways were predicted to be regulated in colostrum EVs compared with mature milk EVs. Notably, Holstein colostrum had the most distinctive miRNA profile, and it was shown that immune-related metabolic pathways were regulated when exposed to human intestinal epithelial cells. It is suggested that analysis of the gene–gene interactions through miRNA profiling and target gene analysis is the cornerstone for screening candidate miRNAs for disease amelioration and identifying the potential of EVs.
















