INTRODUCTION
Coloration is a very important phenotypic trait with various functions related to environmental adaptation, such as temperature regulation and protection against sunburn [1–3], as well as mimicry or camouflage [2–4]. There has long been interest in studying the pigments influencing plumage color in birds, coat color in mammals, and skin color in humans [5]. The plumage color in chickens is genetically complex compared to the coat and skin color in mammals and humans, respectively [5,6]. Besides adapting to the environmental conditions, the plumage color in birds is also an economic trait in the poultry industry with producers and consumers preferring birds of a particular color. For example, the producers of broilers prefer white birds for ease of cleaning and a uniform appearance [7] as well as the easy removal of the feathers [8]. From the consumer’s perspective, plumage color is favored for religious reasons or nutritional value [3].
Plumage color in birds has a key role in sexual selection [2,3,6,9–11] and parent–offspring communication [11]. Melanin is the major pigment producing color in birds and other animals, followed by carotenoids [5,8,11]. Plants, bacteria, and fungi can synthesize carotenoids, while birds and other animals must obtain this pigment from their diet [2,5,11,12]. Carotenoids produce orange, red, and yellow colors in the plumage, bill, skin, and iris [11]. Other pigments such as porphyrins and polyenes influence plumage color in birds [5,6]. Melanin mainly consists of eumelanin and pheomelanin pigments, and eumelanin controls black or brown colors, whereas pheomelanin controls red or yellow colors [3,4,11–14]. Melanin pigments also produce other color patterns such as stripes, spots, and bars in chicken feathers [11,15]. Melanin is produced by melanocytes [13,16]. It is accumulated in melanosomes [3,17] and then transported to keratinocytes [5,10] to give a particular color to the bird’s feathers [5]. In addition to determining color, melanin pigments are associated with antioxidant capacity as well as resistance to bacterial degradation [1,6,11,18].
The production of color in birds is a complex process that is mainly influenced by genetic factors such but also environmental factors [6]. The biosynthesis of melanin depends on tyrosinase activity [9,19,20], and both melanin pigments (eumelanin and pheomelanin) are tyrosine derivatives [4,5]. High tyrosinase activity is associated with the synthesis of eumelanin, whereas low activity results in the production of pheomelanin [4,19,21]. Previous studies have reported that the amount, deposition, distribution, and ratio of melanin pigments affect plumage color in birds [4,5,11,14,20]. Many genes control plumage color in chickens [22]. These genes are involved in melanin synthesis, melanosome transport, melanocyte development, and differentiation [14], and mutations in these genes lead to different colors. For example, the diluted coat color also known as albinism in different species is due to the complete cessation of melanin synthesis caused by mutations in the tyrosinase (TYR) gene and other related genes [9, 14, 23].
Previous studies have explored melanin-related genes (e.g., MC1R, TYR, PMEL; MLPH, ASIP, SLC45A2, EDNRB2, CDKN2A, and SOX10) and confirmed their effects on variation in plumage color in chickens [6,24,25]. The premelanosome protein (PMEL) also known as melanocyte protein Pmel 17 (PMEL17) is encoded by the PMEL gene which is mapped on chromosome 33 and plays a key role in the formation of melanosomes [3,26] and the formation of fibrils on which eumelanin is deposited [5,10,26]. This gene affects the shape of melanosomes [11] and is also involved in the production of eumelanin [10]. Indels in the gene are associated with the dominant white, dun, and smoky colors in chicken plumage [13,27]. A missense mutation in PMEL17 gene is associated with a silver coat color in horses [14,28].
Previous studies have explored the sex-linked barring phenotype in chickens and have reported that the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene located on chromosome Z is responsible for barred plumage in chickens [3,13,25,29]. The four mutations in the CDKN2A gene cause a higher expression of CDKN2A, resulting in a reduction of melanoblasts [3,5,25], thus causing a white bar to appear where melanocytes are absent [5,25]. The methylthioadenosine phosphorylase (MTAP) gene is mapped on chromosome Z and acts as an inhibitor of dermal melanin in chickens and as a tumor suppressor in humans, thus inhibiting melanoma cell proliferation [3]. It is also thought to be involved in the barring plumage of chickens [29]. In the chicken genome assembly (GRCg6a), the PMEL gene is accessed by ENSGALG00000035350 whereas CDKN2A and MTAP genes can be accessed by ENSGALG00000034505 and ENSGALG00000008174, respectively.
A previous genome-wide association study (GWAS) of plumage colors have reported three potential candidate genes that could affect the variation in plumage color in chickens, including the CDKN2A, PMEL, and MTAP genes [3]. However, the genotype effect of these genes on the variation in chicken plumage color are not fully understood. Therefore, we investigated single-nucleotide polymorphisms (SNPs) in the CDKN2A, PMEL, and MTAP genes to assess their effects on the variation in plumage color in Yeonsan Ogye-White Leghorn crossbred chicken’s population.
MATERIALS AND METHODS
The Animal Ethics Committee of Chungnam National University (no. 202103A-CNU-061) approved this study to abide by the standard guidelines for animal care.
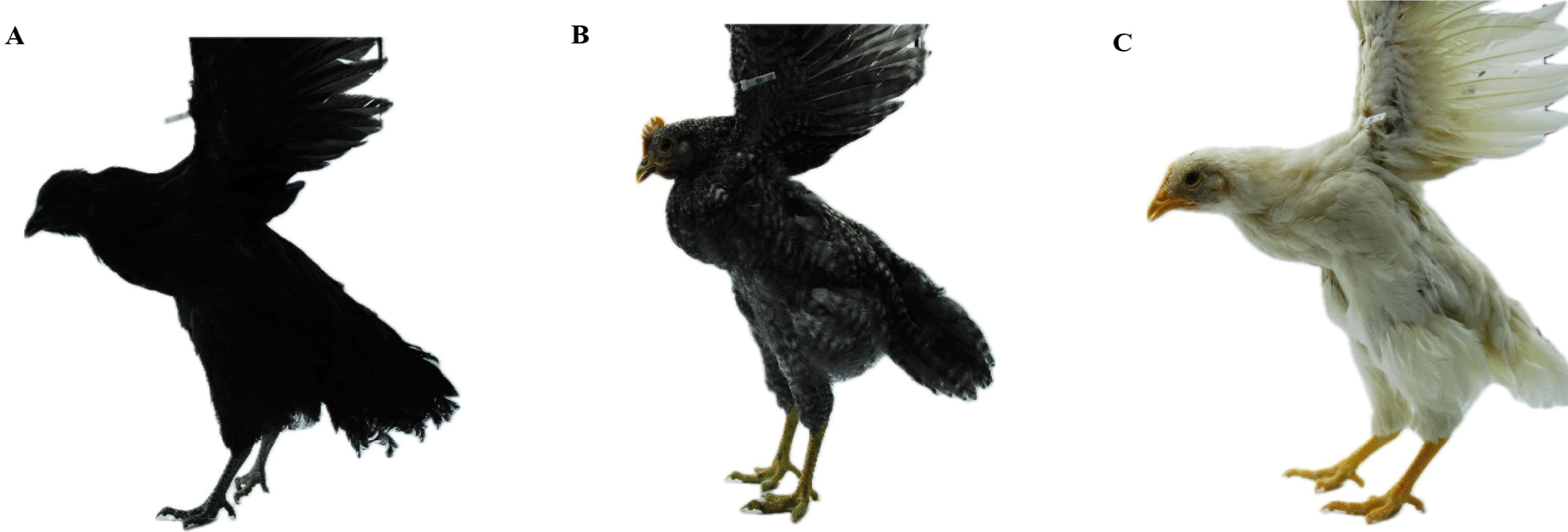
This study used a total sample of 428 birds collected from an F2 population between Yeonsan Ogye and White Leghorn with the three plumage color phenotypes: all black, n = 62, all white, n = 246, and black and white barred (barred), n = 120 as shown in Fig. 1. Yeonsan Ogye is a Korean native chicken breed that is completely black from beak to toes [3,8,30] as well as bones and internal organs [3]. For phenotyping, all while the plumage color of Yeonsan Ogye is completely black, the White Leghorn has a completely white plumage color [30]. F2 population was produced by crossing the F1 progenies. The F1 population was produced by crossing Yeonsan Ogye and White Leghorn. Three phenotypes (all-black, all-white, and black and white barred) were selected, the chicken’s photos were taken by the National Institute of Animal Science (NIAS) in 2020 using a digital camera (D80, Nikon, Tokyo, Japan) as described in [3].
The chickens used in this study were kept under the same management conditions at the Animal Genetic Resources Research Center’s farm at the NIAS, Korea. Genomic DNA was extracted from blood samples of birds at 8 weeks of age using the Wizard Genomic DNA Purification Kit (Promega). DNA stocks were diluted with deionized distilled water to produce a working concentration of 25 ng/μL and stored at −20°C.
Two pairs of primers were designed to amplify the fragment of 440 bp and 293 bp for missense variant: rs14684281T/C and missense variant: rs312616138A/G in the PMEL gene, respectively. A fragment of 351 bp and a fragment of 623 bp were also amplified to identify the SNPs: synonymous variant: rs316391660C/T and a missense variant: rs1058656732C/T in the MTAP and CDKN2A genes, respectively. We designed these primers by primer-BLAST tool and were synthesized by Bioneer. The primers used in this study are presented in Table 1. We performed the PCR amplification using the same conditions as described in our previous work [31]. Annealing temperatures for each primer set are presented in Table 1.
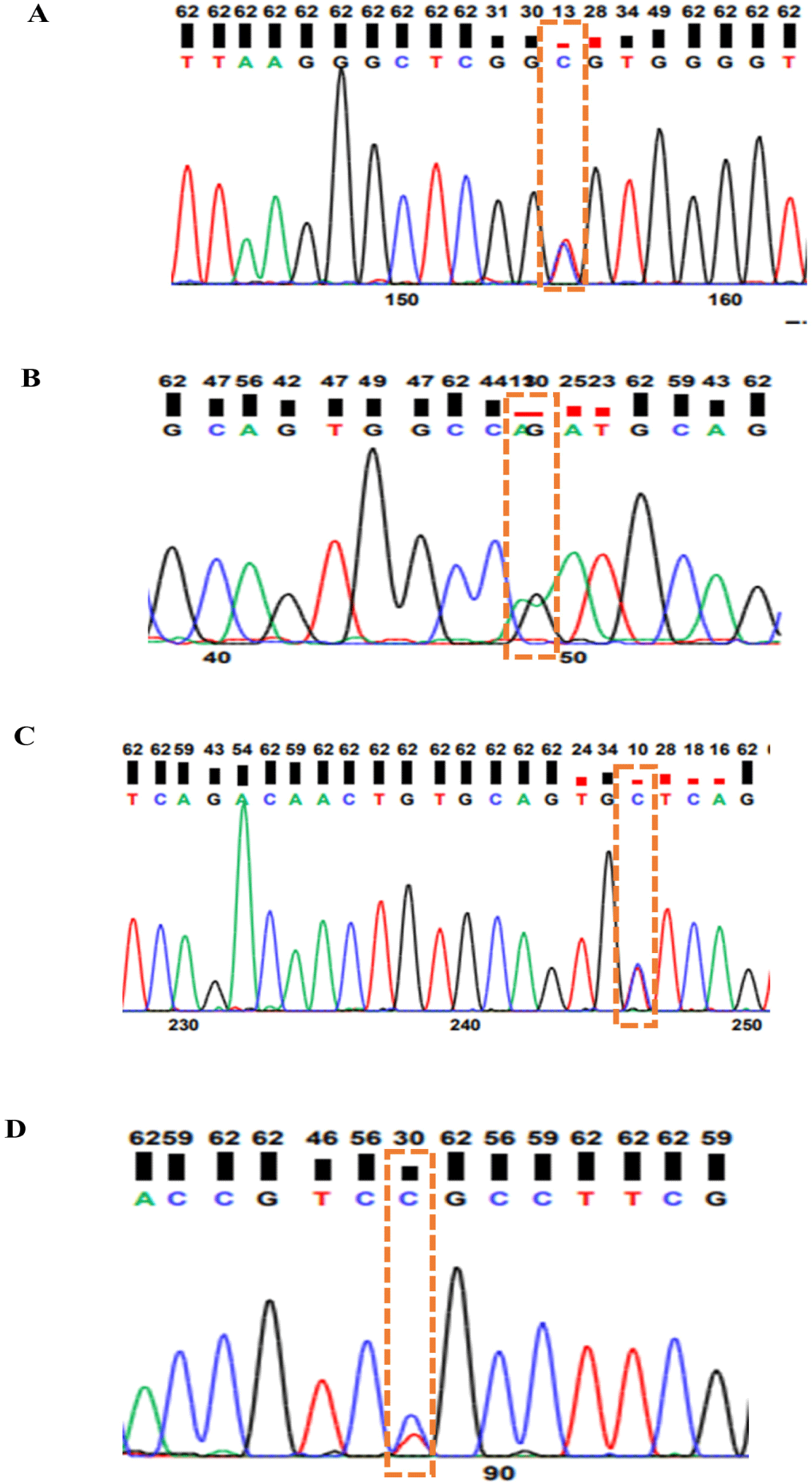
Before sequencing, the PrimePrep PCR Purification Kit (GenetBio) was used to purify the PCR products, and spectrophotometry (NanoDrop 2000, Thermo Fisher Scientific) was used to check the quality of DNA. Sequencing was performed by Bioneer . One missense variant was confirmed in the CDKN2A gene (rs1058656732C/T), and in the MTAP gene, one synonymous variant was identified (rs316391660C/T) whereas two missense variants were found in the PMEL gene (rs14684281T/C and rs312616138A/G) (Fig. 2).
The PCR allelic competitive extension (PACE) technology was used for genotyping our targeted SNPs. We prepared the SNP target-specific primers for the PACE genotyping assay (Table 2). The PACE assay mix and PACE master mix are shown in Table 2. The PACE assay mix and PACE master mix were synthesized by 3CR Bioscience. A 96-well plate was used for genotyping, and each well had 10 μL made up of 1 μL of genomic DNA (5 ng/μL) or 1 μL of 3DW for negative control, 5 μL of master mix, 0.25 μL of assay mix, and 3.75 μL of 3DW. We run the reaction by using the CFX ConnectTM Real-time PCR Detection System (Bio-Rad Laboratories).
Association analysis between the genotypes of MTAP gene (rs316391660C/T) and PMEL gene (rs312616138A/G and rs14684281T/C) with plumage color variations in Yeonsan-Ogye-White leghorn crossbred chickens was performed by Fisher’s exact test. Fisher’s exact test is appropriate for small sample size or if some expected frequencies are less than 5. All calculations were carried out by using the R program [32]. A significant association was confirmed when p < 0.05.
RESULTS
Sequencing the target genes was performed to detect different variants in the F2 population. Two missense variants were detected in PMEL (rs14684281T/C, rs312616138A/G); one synonymous mutation was detected in MTAP (rs316391660C/T), and one missense mutation was detected in CDKN2A (rs1058656732C/T) as shown in Fig. 2. A missense SNP: rs14684281T/C of the PMEL gene is located in exon 2 whereas a missense SNP: rs312616138A/G is located in exon 6 of the PMEL gene mapped on chromosome 33. Moreover, a missense SNP: rs1058656732C/T of the CDKN2A gene is found in exon 1 whereas a synonymous SNP: rs316391660C/T is found in exon 6 of the MTAP gene. Both CDKN2A and MTAP gene are mapped on Z chromosome. All variants detected by sequencing in the CDKN2A, PMEL, and MTAP genes were genotyped by the PACE genotyping method.
PACE genotyping result of the CDKN2A, MTAP, and PMEL genes showed that the rs1058656732C/T variant in CDKN2A gene has one CC genotype in all F2 population, which means that the variant is monomorphic in the F2 population. Two missense variants (rs312616138A/G; rs14684281T/C) in the PMEL gene resulted in three genotypes each (AA, AG, and GG; CC, CT, and TT, respectively; Fig. 3), and a synonymous SNP (rs316391660C/T) in the MTAP gene resulted in three genotypes: CC, CT, and TT (Fig. 3).
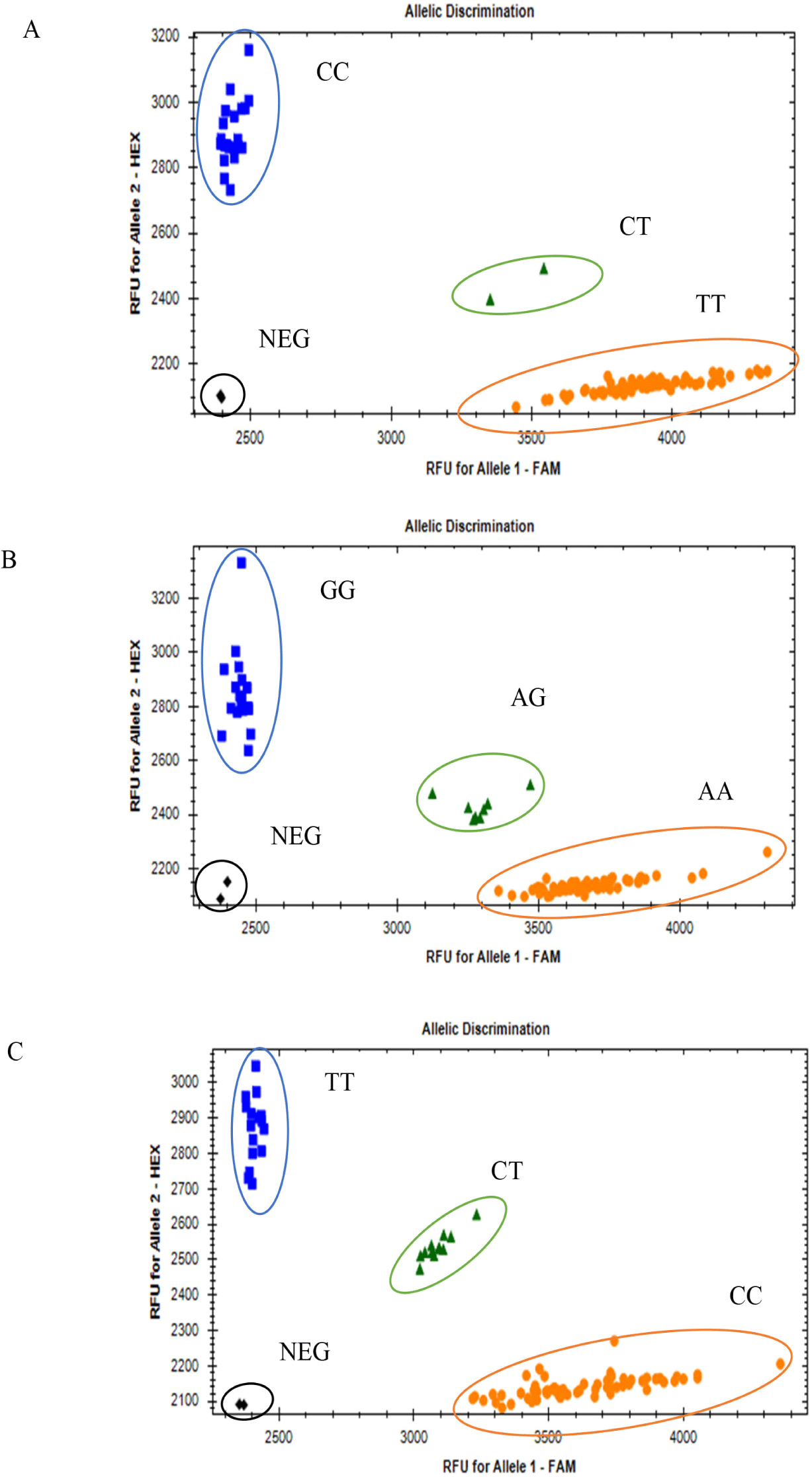
Regarding the synonymous SNP (rs316391660C/T) of the MTAP gene, the TT genotype had the highest frequency (52.1%), followed by the CT (24.4%) and CC genotypes (23.5%) in a total population of 428 chickens, consisting of all-black (62), all-white (246), and black-and-white barred (120) chickens (Table 3). Regarding one missense SNP of PMEL (rs312616138A/G), the homozygous AA genotype had the highest frequency (50.2%), followed by the GG genotype (46.5%) and heterozygous AG genotype (3.3%) (Table 3). Regarding the other missense SNP (rs14684281T/C), the homozygous genotype CC had the highest frequency (58.9%), followed by the homozygous TT genotype (26.2%), and the heterozygous CT genotype had the lowest frequency (14.9%) (Table 3). The genotype and allele frequencies in each class of plumage color are shown in Table 3.
To evaluate the MTAP and PMEL genotype effects on the variation in plumage color, we performed the chi-square and Fisher’s exact tests. The MTAP (rs316391660C/T) and PMEL genotypes (rs312616138A/G and rs14684281T/C) had a significant (p < 0.05) influence on the three plumage color variants (Table 3). For rs316391660C/T variant of the MTAP gene, the frequency of the homozygous CC genotype in all-black chickens was higher than in white and black-and-white barred chickens, whereas the frequency of the homozygous TT genotype was higher in all-white chickens than in all-black and black-and-white barred chickens. For the rs312616138A/G missense SNP of PMEL, the homozygous AA genotype had a greater frequency in all-white and black-and-white barred chickens compared to that in all-black chickens, while the frequency of the GG genotype was higher in all-white chickens. For the rs14684281T/C missense SNP of the PMEL gene, the frequency of the TT homozygous genotype was higher in black-and-white barred chickens than in all-black and all-white chickens, whereas the homozygous CC genotype had the highest frequency in all-white chickens compared to all-black and black and white chickens. Due to sexual dimorphism for plumage color traits, association test between the genotypes and plumage color variations was performed separately in males and females, and results are shown in Table 3.
DISCUSSION
We explored one variant (rs316391660C/T) in the MTAP gene, one (rs1058656732C/T) in the CDKN2A gene, and two (rs312616138A/G; rs14684281T/C) in the PMEL gene to confirm their genotype effects on the coloration of chicken plumage which is influenced by many genes [22]. These genes have been reported to be involved in the synthesis of melanin, melanosome transport, melanocyte development, as well as their differentiation [14]. In previous study, it has been reported that the PMEL gene is associated with the dominant white, dun, and smoky colors in chicken plumage [27]. The insertion of 9 bp in exon 10 of PMEL gene inhibits the synthesis of eumelanin in feathers [3,27]. Another candidate gene MTAP causes the barring plumage of chickens [29]. The white bar that appears in black-and-white barred plumage is formed due to the absence of melanocytes [25]. In this study, we explored the influence of these two genes and their effects on three plumage colors: all-black, all-white, and black-and-white barred chickens. We report a significant influence of both of these genes. This study found two missense variants in the PMEL gene. A SNP: rs14684281T/C found in exon 2 lead to a change of amino acid from valine (V) to alanine (A) at the 35th position of the protein whereas a SNP: rs312616138A/G in exon 6 and lead to a change of amino acid from asparagine (N) to aspartic acid (D) at the 399th position of protein. Furthermore, we checked the sorting intolerant from tolerant (SIFT) scores for two SNPs (rs14684281T/C and rs312616138A/G) in PMEL gene and were likely to be tolerated (0.95 and 0.71, respectively) which means the change of amino acid does not significantly affect the protein function. The change for amino acid at the beginning of the protein for the SNP: rs14684281T/C (V35A in exon) and that of SNP: rs312616138A/G (N399D) might affect the protein folding or protein stability thus affecting the formation of melanosome.
To the best of our knowledge, this is the first study to report the association between the MTAP genotypes and the plumage coloration in chickens. For rs316391660C/T variant in the MTAP gene, the heterozygous CT and homozygous TT genotypes were absent in all-black chicken population (Table 3). The homozygous CC genotype was more frequent in all-black chickens and the allele C was fixed (100%) in all-black chickens (Table 3). This fixation was probably due to the artificial selection [33]. In this study, we used F2 population between Yeonsan Ogye and White Leghorn. The F0 population was sampled from a small population of Yeonsan Ogye, which may contribute to the fixation of the C allele in rs316391660 C/T (synonymous) locus of the MTAP gene in the all-black chickens. For rs14684281T/C variant in the PMEL gene, the homozygous CC genotype was more frequent than the CT and TT genotypes in all-white chicken population. All three genotypes (CC, CT, and TT) were significantly associated with all three plumage colors: all-black, all-white, and black-and-white barred chickens.
The plumage color bas been known to be a complex trait influenced by several genes. We discovered that one synonymous SNP (rs316391660C/T) in the MTAP and two missense SNPs (rs312616138A/G and rs14684281T/C) in the PMEL genes have a significant genotype effect on the three plumage colorations. These results confirm the genotype effects of the PMEL gene on the dominant white plumage color, and suggest that the MTAP gene could be used as a genetic marker for the breeding of chickens with black-and-white barred plumage. However, further studies exploring the biological functions of the MTAP gene and identifying different variants using large-scale sample sizes are needed.
CONCLUSION
In this study, we detected the SNPs in the CDKN2A, PMEL, and MTAP genes and assess their genotype effects on the variation in plumage color in chickens. Our results showed that the synonymous SNP (rs316391660C/T) in the MTAP gene and two missense SNPs (rs312616138A/G; rs14684281T/C) in the PMEL gene were found to have a significant genotype effect on plumage color in Yeonsan Ogye-White Leghorn crossbred chicken population. Our findings could help to elucidate the genetic mechanisms underlying chicken plumage coloration. Furthermore, the synonymous SNP (rs316391660C/T) in the MTAP gene can be used as a genetic marker for breeding chickens with black-and-white barred plumage.
















