INTRODUCTION
The fertilization of an oocyte with a spermatozoon is followed by a period of transcriptional quiescence of varying lengths when the embryonic genome contained in the nucleus is not yet expressed. During this period of transcriptional silencing, development is driven by cytoplasmic factors that mainly consist of maternally deposited mRNA [1]. To ensure this developmental process, maternal mRNA activity is restricted to a precise time and space [1]. Translation is a major regulatory step in the translation of maternally stored mRNAs [2], suggesting that translation and post-translational regulation may play key roles in early embryogenesis. The maternal-to-zygotic transition (MZT) process includes zygotic genome activation (ZGA) [3]. According to previous studies, the major embryonic transcriptional activation of porcine ZGA occurs at the four-cell stage [4–6]. Moreover, the also regulation of genomic transcriptional activity during early embryonic development is governed by histone modifications, including processes like lysine acetylation and lysine methylation [7]. ZGA failure will causes abnormal embryonic development [4,8].
Metabolic programming is closely related to early embryonic development, including ZGA. Mitochondria are one of the most important organelles in the cell and are responsible for metabolic, generating energy and participating in various physiological processes such as apoptosis [9, 10]. Mitochondrial tricarboxylic acid cycle enzymes, which are normally located in the mitochondria, are also important for mouse ZGA [11]. During the two-cell stage, zygotic cells upregulated the expression of genes for pyruvate metabolism in mitochondria and oxidative phosphorylation [12]. Therefore, the mitochondria is crucial for ZGA.
Y-box-binding protein 2 (YBX2), alternatively referred to as MSY2, is a germ cell-specific Y-box-binding protein that is expressed in germ cells of the adult testis as well as in the developing fetal testis and ovary, but not in other normal tissues [13–15]. YBX2 is important for round spermatids because it represses translational activity and transcript degradation [16], suggesting that YBX2 plays a role in regulating the translation of paternal mRNAs. Additionally, YBX2, which is specifically expressed in oocytes, is the most abundant RNA-binding protein (approximately 2% of total oocyte protein), and it is degraded at the 2-cell stage, which corresponds to the degradation cycle of maternal mRNAs in mouse [17]. Furthermore, the knockout/knockdown of the YBX2 can lead to a comprehensive reduction in mRNA within developing oocytes, thereby causing issues with oocyte maturation and early embryonic development [18, 19]. And, it has been shown that YBX2 stabilizes oocyte mRNA through reversible spongy cortical partitioning-dependent [20]. Thus, these findings suggest that YBX2 plays a critical role in storing and stabilizing maternal mRNA and early embryonic development. Moreover, YBX2 stabilizes mRNA targets that encode proteins involved in mitochondrial function [21]. Based on this information, we investigated the impact of YBX2 on early porcine embryonic development.
In this study, the effects of YBX2 on the mitochondria, mRNA translation, and gene transcription were investigated using siRNA microinjection at the single-cell stage. These findings indicate that YBX2 plays a role in mitochondrial function and ZGA.
MATERIALS AND METHODS
Unless specified otherwise, all chemicals were sourced from Sigma-Aldrich.
Ovaries from prepubertal gilts were obtained from a local abattoir (Farm Story Dodarm B&F) and rinsed three times in saline solution containing 75 mg/mL of penicillin G and 50 mg/mL of streptomycin sulfate at 37°C. Follicles approximately 3–6 mm in diameter were aspirated using a 10-mL disposable syringe. Selection criteria for the cumulus-oocyte complexes (COCs) included a minimum of three layers of compact cumulus cells. After three rinses with in vitro maturation (IVM) medium (TCM-199 [11150-059, Gibco] supplemented with 100 mg/L sodium pyruvate, 10 ng/mL epidermal growth factor, 10% [v/v] FF, 10 IU/mL LH, and 10 IU/mL follicle stimulating hormone), approximately 70 COCs were were placed into 4-well plates with 500 μL of IVM medium covered by mineral oil and cultured for 44 h at a temperature of 38.5°C in an atmosphere containing 5% CO2.
The COCs were treated with 1 mg/mL hyaluronidase and pipetted approximately 50 times until the oocytes were naked and oocytes with the first polar bodies were selected. Next, two pulsed direct current of 110 V for 60 μs was applied to parthenogenetic activation of MII oocytes in 0.1 mM CaCl2, 0.05 mM MgSO4, 0.01% polyvinyl alcohol (PVA; w/v), and 0.5 mM hydroxyethyl piperazine Ethane Sulfonicacid. The activated oocytes were cultured in porcine zygote medium 5 (PZM-5) with 4 mg/mL bovine serum albumin (BSA) and 7.5 μg/mL cytochalasin B for 3 h. Subsequently, the oocytes underwent thorough washing and were then incubated in PZM-5 medium added with 0.4% BSA for 6 days at 38.5°C (5% CO2). The blastocyst (BL) rate was determined on day 6.
For knockdown (KD) experiments, small interfering RNAs (siRNAs) were were crafted to target three distinct regions within the porcine genome (GenePharma). All siRNA sequences used in this study are listed in Table 1. A mixture of YBX2-1, YBX2-2, and YBX2-3 siRNAs was prepared for microinjection. YBX2 siRNA (50 μM) was microinjected into the cytoplasm of the zygotes using the Eppendorf Femto-Jet (Eppendorf) and Nikon Diaphot Eclipse TE300 inverted microscope (Nikon,) equipped with the Narishige MM0-202N hydraulic 3-D micromanipulator (Narishige). Following the injection, the embryos were nurtured in PZM-5 medium for either 2 or 6 days.
| siRNA | siRNA sequences |
|---|---|
| siYBX2-1 | F: GUUCACCAGACAGCUAUUATT R: UAAUAGCUGUCUGGUGAACTT |
| siYBX2-2 | F: GAAGCUGCUAACGUAACUGTT R: CAGUUACGUUAGCAGCUUCTT |
| siYBX2-3 | F: GGUAGAACCCAAAGAGACATT R: UGUCUCUUUGGGUUCUACCTT |
Embryos were fixed using a 3.7% solution of paraformaldehyde (PFA) for 30 min at room temperature (RT), followed by three washing with PVA/phosphate buffered saline (PBS). Subsequently, they were permeabilized with 1% Triton X-100 for 30 min and blocked with a solution of 3.0% BSA with 0.1% Triton X-100 for 1 hour at RT. Embryos were incubated overnight at 4°C with different primary antibodies. After washing three times with PVA/PBS, the embryos were treated with different second antibodies (1:200; A10040, Invitrogen) for 1 h at RT. The embryos were then mounted onto slides using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories) and visualized using a confocal microscope (Zeiss LSM 710 META, Zeiss). The resulting images were processed with Zen software version 8.0 (Zeiss).
Embryos were treated with MitoTracker Red CMXRos at a concentration of 500 nM (M7512, Invitrogen) for a duration of 30 min at a temperature of 38.5°C. Post-incubation, they were washed thrice with PVA/PBS. Subsequently, the embryos were fixed in 3.7% PFA for 30 min at RT and washed an additional three times with PVA/PBS. Finally, the embryos were mounted onto slides.
mRNA was isolated from 30 embryos from the control and YBX2 KD groups respectively, utilizing the DynaBeads mRNA Direct Kit (61012, Thermo Fisher Scientific), following the protocol provided by the manufacturer. The RNA was converted into cDNA using oligo (dT) 20 primers and SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). For real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR), the WizPure qPCR Master Kit was utilized. A 20 μL reaction mixture was prepared, consisting of 10 μL SYBR Green, 1 μL each of the forward and reverse primers, 2 μL of cDNA, and 7 μL of double distilled water (ddH2O). The amplification cycle was set as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 60°C for 25 s, 72°C for 10 s, and a final extension at 72°C for 5 min. The 18S rRNA gene served as reference gene. The primer sequences for each target gene are detailed in Table 2. mRNA quantification was calculated using the 2−ΔΔCt method.
Sixty embryos each from the control and YBX2 KD groups were combined with 20 μL of ice‐cold Laemmli sample buffer (sodium dodecyl sulfate [SDS] sample buffer that includes 2‐mercaptoethanol) and heated at 95°C for 10 min. The proteins from each sample were then separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore). To prevent nonspecific binding, the membranes were blocked with Tris-buffered saline with Tween-20 (TBST), supplemented with either 5% skim milk powder or BSA, for 1 h at RT. Subsequently, the membranes were incubated with different primary antibodies in a blocking solution overnight at 4°C. After washing three times with TBST (10 min each), the membranes were treated with secondary antibodies (1:20000) for 1 h at RT. The membranes were subsequently developed using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). The resulting band intensities were quantified using ImageJ software.
Each experiment was performed at least in triplicate. Data were analyzed using the GraphPad Prism 5 software (GraphPad). Statistical analysis was performed by t-test between control and YBX2 KD group. All data are shown as the mean ± SEM. Statistical significance was set at p < 0.05.
RESULTS
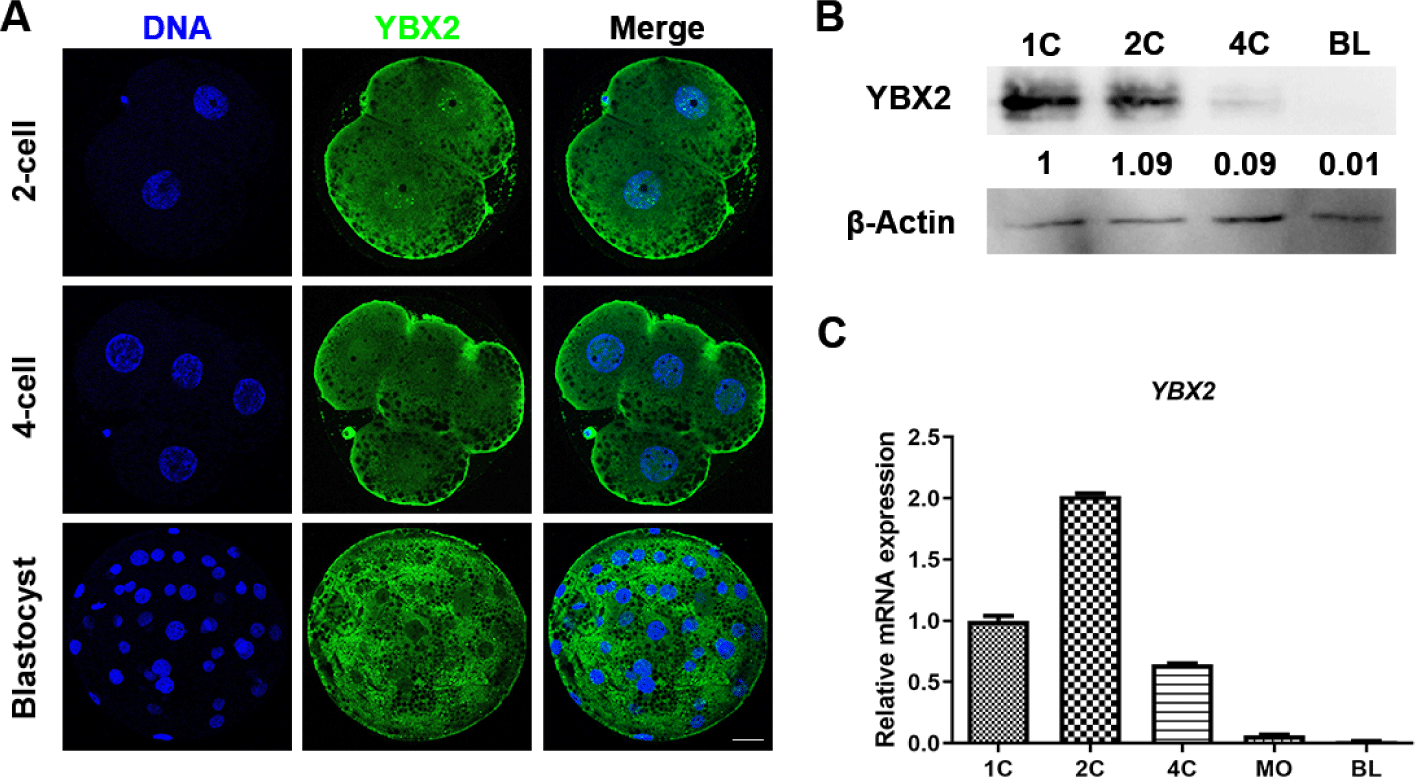
To ascertain the subcellular distribution of YBX2 during embryonic development, immunofluorescence staining was conducted to delineate its location in two-cell (2C; n = 10), four-cell (4C; n = 10), and BL (n = 10) embryos. As shown in Fig. 1A, YBX2 is localized in the nucleus, cytoplasm and cortex during the 2C and 4C stages. However, it was localized in the cytoplasm at the BL stage. Next, we determined the protein expression levels of YBX2 by western blotting, the result indicated that YBX2 was present during porcine embryonic development (Fig. 1B). Subsequently, using RT-qPCR, we observed that the mRNA levels of YBX2 were the highest at the 2C stage and decreased at the 4C stage (Fig. 1C).
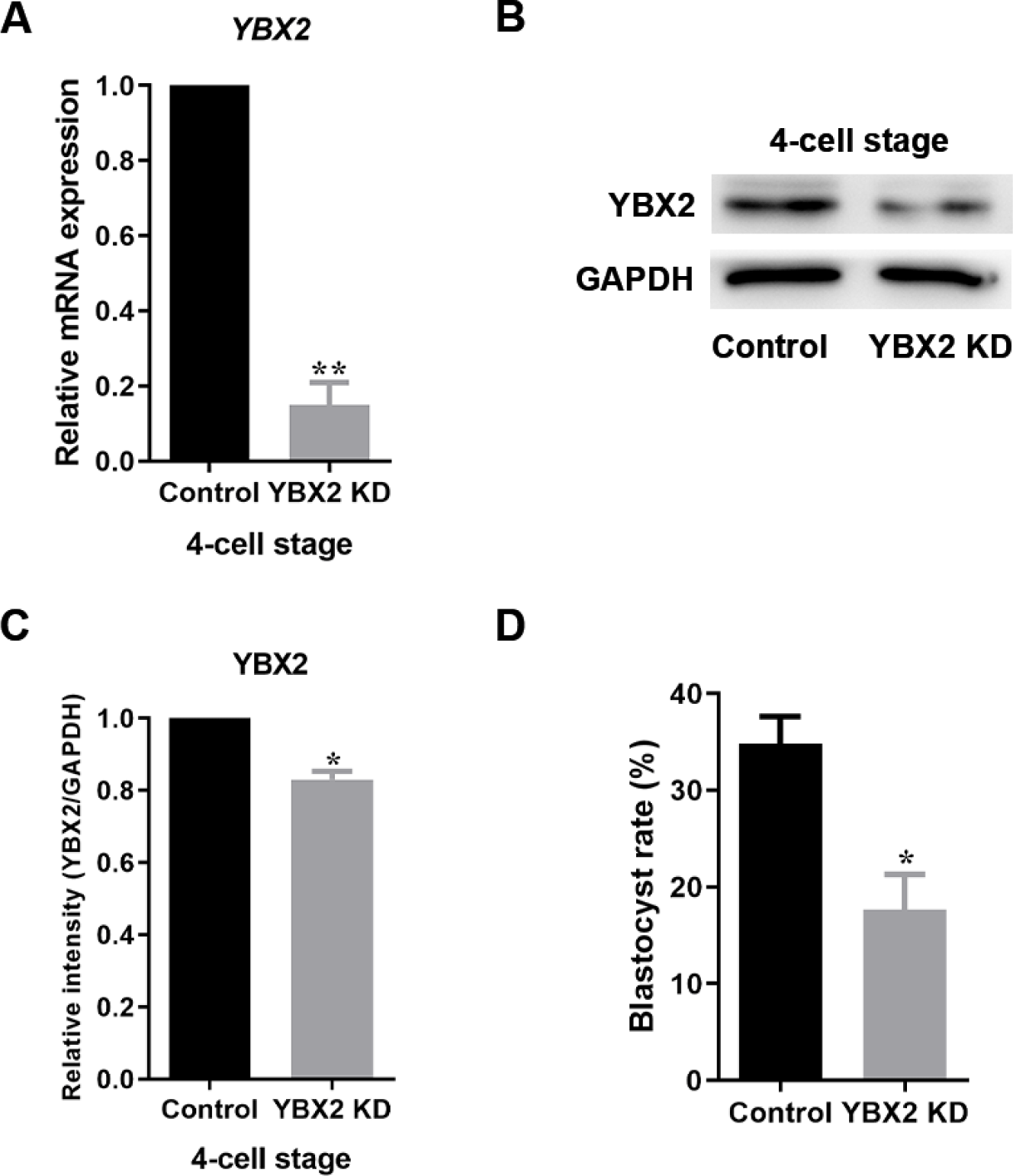
To study the effects of YBX2 on porcine embryonic development, YBX2 (YBX2 KD) siRNA was microinjected into the zygote. YBX2 KD resulted in a significant decrease in mRNA levels at the 4C stage (1 vs. 0.15 ± 0.06; p < 0.01; Fig. 2A). Western blotting also demonstrated that YBX2 protein expression levels were reduced (1 vs. 0.83 ± 0.02; p < 0.05; Figs. 2B and 2C). Upon YBX2 KD, the BL formation rate in the YBX2 KD group was significantly lower than that in the control group (34.81 ± 2.86 vs. 17.74 ± 3.69; p < 0.05; Fig. 2D), indicating that YBX2 plays an important role in embryo development.
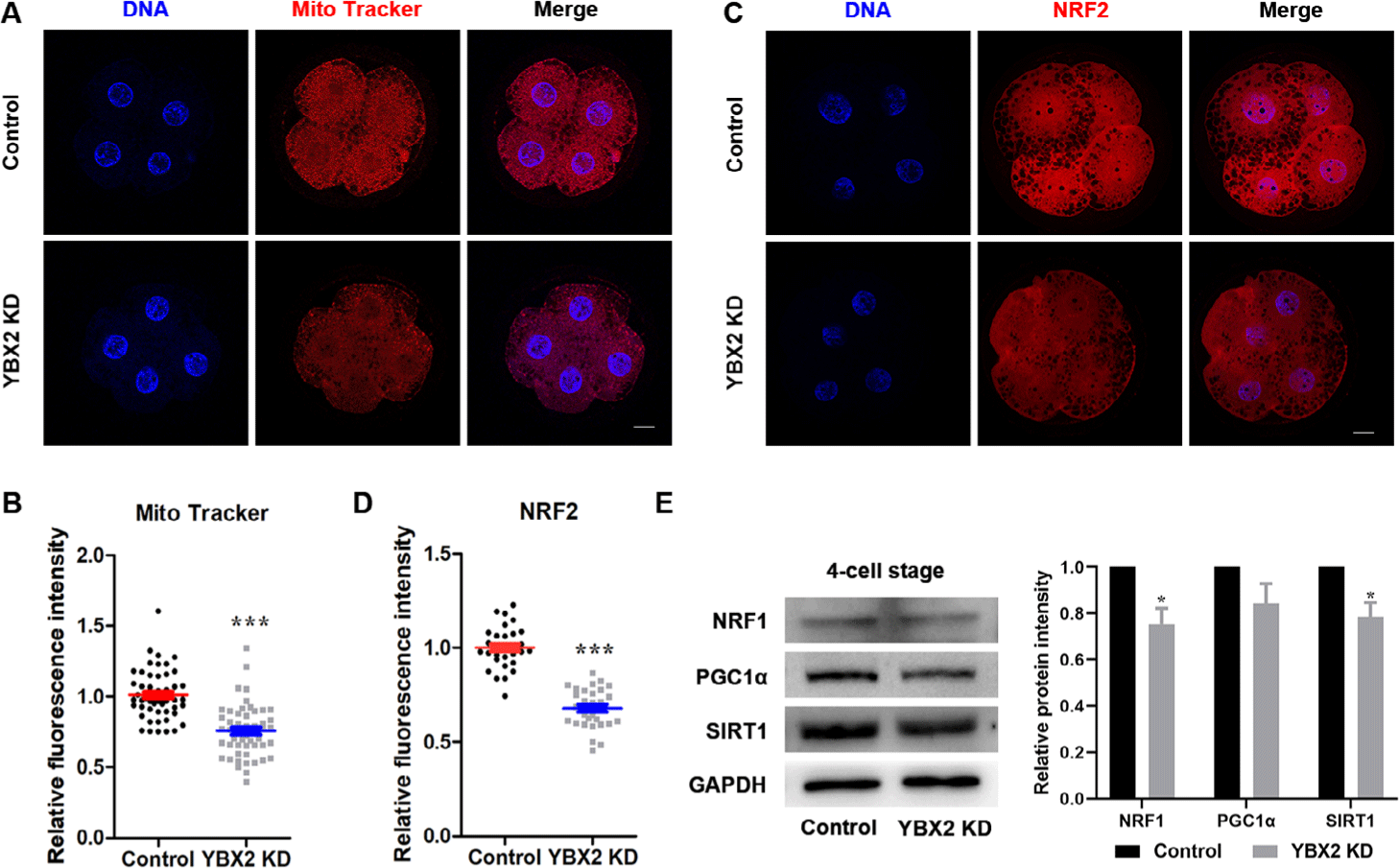
YBX2 stabilizes mRNA targets that encode proteins rich in mitochondrial function. To determine mitochondrial activity, embryos were stained with MitoTracker Red CMXRos at the 4C stage. As shown in Figs. 3A and 3B, the intensity of active mitochondria was significantly decreased after YBX2 KD (1.01 ± 0.02 vs. 0.76 ± 0.03; p < 0.001). PGC1α, SIRT1, NRF1, and NRF2 impact mitochondrial biogenesis. At the 4C stage, immunofluorescence staining revealed that NRF2 expression levels were significantly reduced upon YBX2 KD (1 ± 0.02 vs. 0.68 ± 0.02; p < 0.001; Figs. 3C and 3D). Western blotting revealed a reduction in the protein levels of SIRT1 (1 vs. 0.79 ± 0.04; p < 0.05; Fig. 3E) and NRF1 (1 vs. 0.75 ± 0.04; p < 0.05; Fig. 3E) upon YBX2 KD. Taken together, these results indicate that YBX2 KD impairs mitochondrial biogenesis.
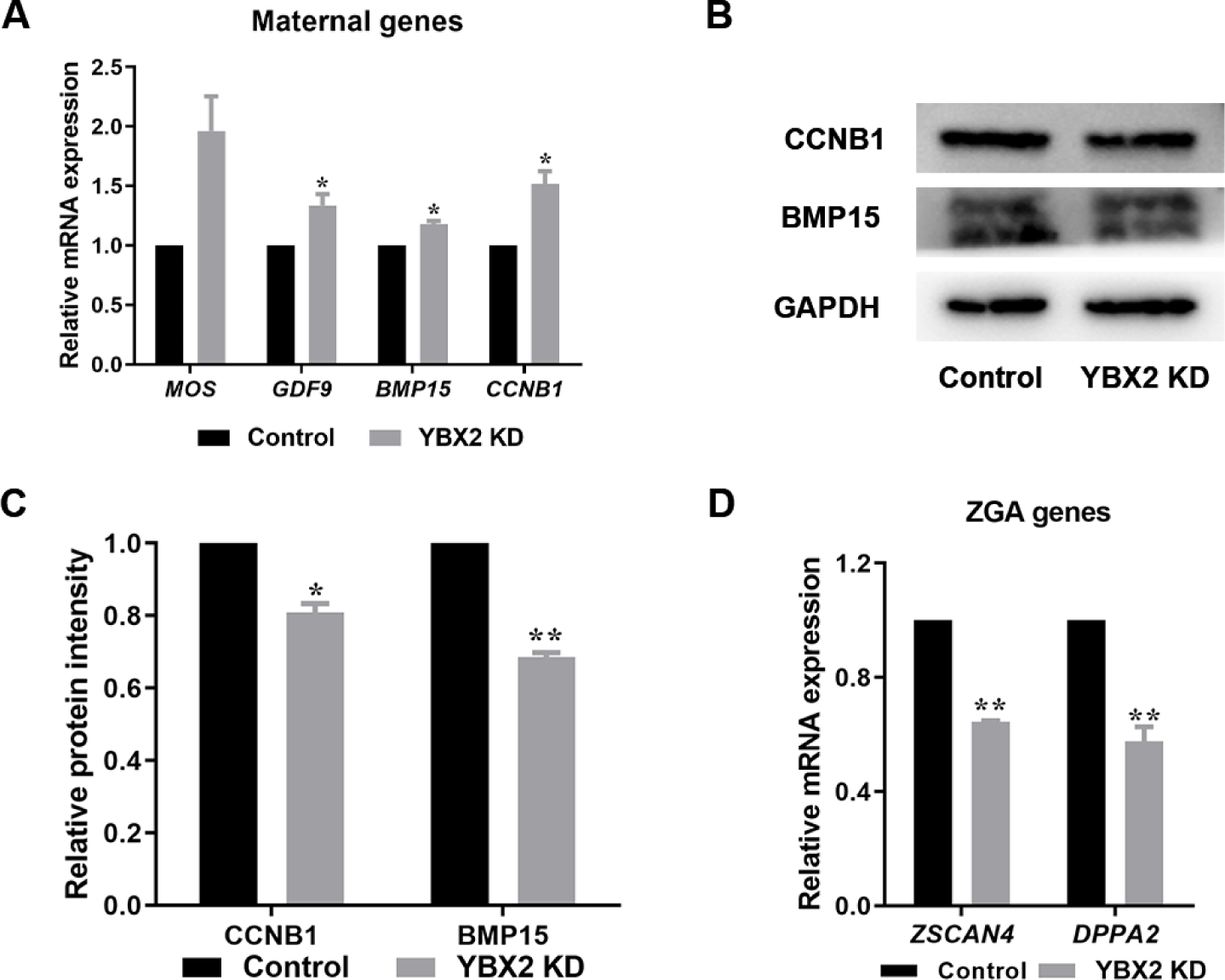
Mitochondrial dysfunction can affect ZGA during early embryonic development [3,22]. Therefore, we investigated the effects of YBX2 on ZGA. Initially, we detected maternal mRNA degradation and ZGA gene expression using RT-qPCR in the control and YBX2 KD groups. As shown in Fig. 4A, the maternal expression levels of GDF9 (p < 0.05), BMP15 (p < 0.05), and CCNB1 (p < 0.05) in the YBX2 KD group was significantly higher than those in the control group. However, there was no significant difference in MOS mRNA levels between YBX2 KD and control groups. In addition, the protein levels of CCNB1 (0.81 ± 0.02 vs. 1; p < 0.05; Figs. 4B and 4C) and BMP15 (0.69 ± 0.01 vs. 1; p < 0.01; Figs. 4B and 4C) were significantly lower in the YBX2 KD group compared to the control group. ZGA genes ZSCAN4 (0.64 ± 0.01 vs. 1; p < 0.01; Fig. 4D) and DPPA2 (0.58 ± 0.05 vs. 1; p < 0.01; Fig. 4D) expression levels were significantly decreased after YBX2 KD. These data indicate that YBX2 KD impairs the ZGA process.
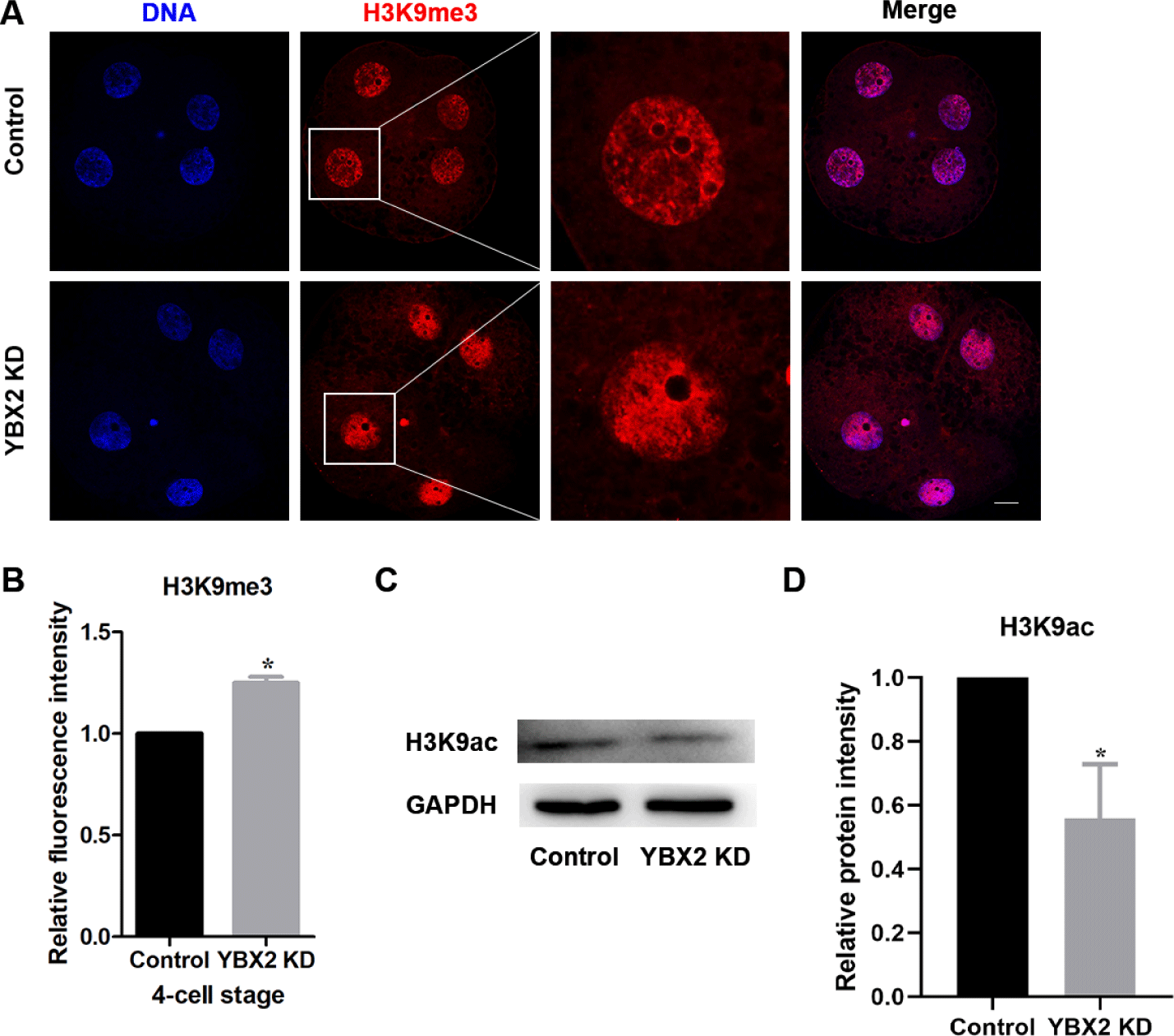
Histone modifications occur throughout the early embryonic development and affect the interactions between transcriptional factors and chromatin. Any irregularities in histone modifications may cause developmental abnormalities in embryos. Therefore, we examined the effect of YBX2 on H3K9ac and H3K9me3 levels. The result showed that the levels of H3K9me3 in the YBX2 KD group was significantly higher than those in the control group (1 ± 0.00 vs. 1.25 ± 0.03; p < 0.05; Figs. 5A and 5B). Moreover, western blotting for H3K9ac was also decreased in the YBX2 KD group (1 vs. 0.56 ± 0.10; p < 0.05; Figs. 5C and 5D). These results indicate that YBX2 affects gene transcription.
DISCUSSION
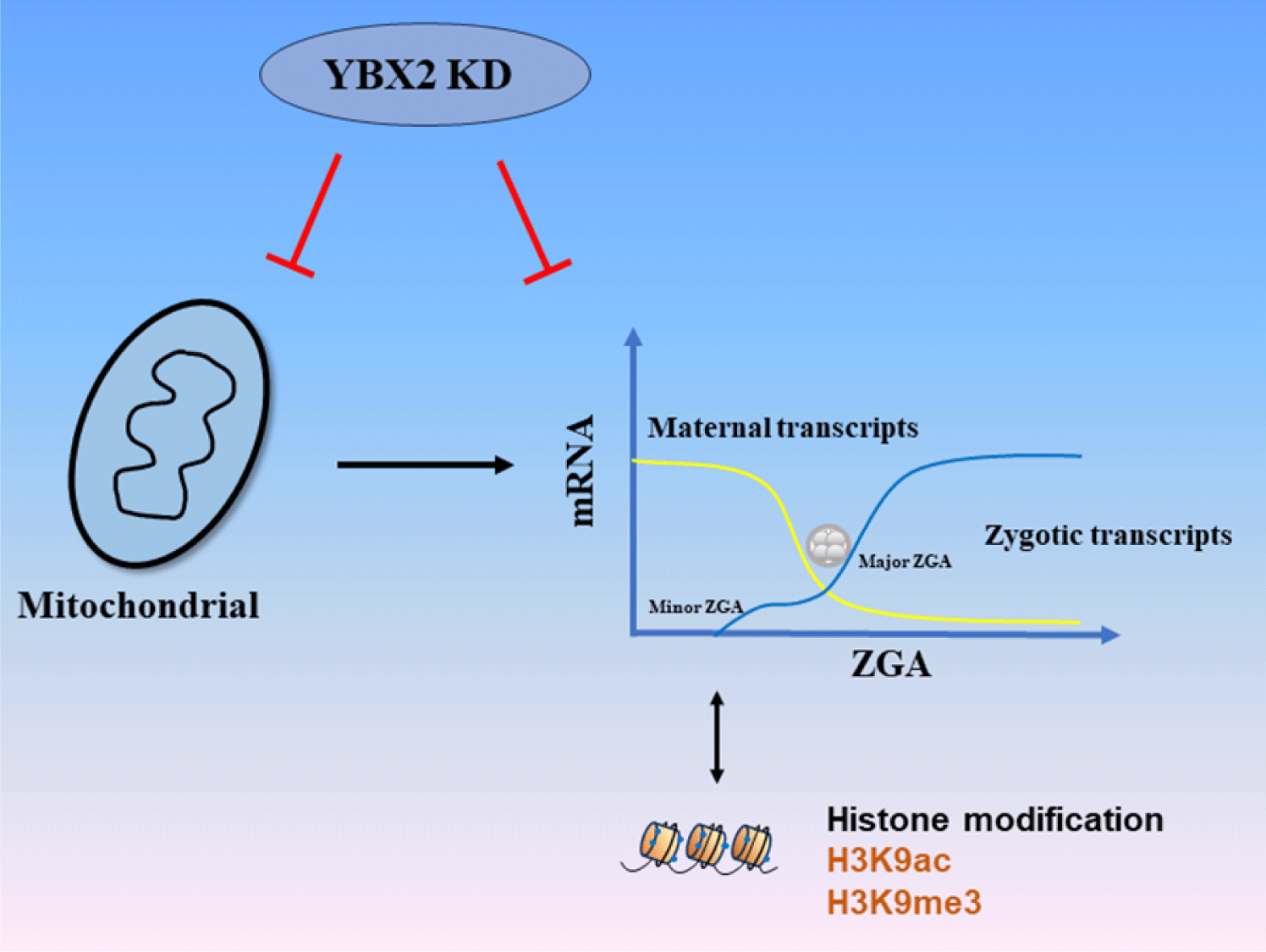
We investigated the function of YBX2 in early porcine embryonic development (Fig. 6). These findings indicated that YBX2 is a maternal gene with important roles in mitochondrial biogenesis and ZGA in porcine embryos. YBX2 KD resulted in mitochondrial dysfunction and anomalies in the ZGA process, leading to abnormal porcine embryonic development.
Bovine embryos revealed that the protein expression levels of YBX2 are analogous to that in mice, with higher abundance at the early cleavage stage and a subsequent decline after the MZT [17, 23], suggesting that YBX2 is prevalent in early embryos. In this study, YBX2 mRNA and protein expression patterns similar to those in bovine were observed in porcine embryos. Depending on its expression profile, YBX2 prevents premature translation of mRNAs in early embryos. Since porcine embryos at the morula stage and bovine embryos at the 16-cell stage can transcribe their own mRNAs they do not require YBX2, suggesting that YBX2 is a maternal gene that plays a crucial role before the start of embryonic transcription. In addition, lack of YBX2 in mouse oocytes interferes with oocyte growth and maturation, RNA stability, and the transcriptome, resulting in reduced fertility [18,19]. In the current study, YBX2 KD decreased the BL formation rate, suggesting that YBX2 plays a crucial role in embryo development. Furthermore, numerous studies have shown that deletion of YBX2 leads to spermatogenic arrest [24,25]. In summary, YBX2 is important in oocyte, sperm, and early embryonic development.
YBX2 stabilizes mRNA targets encoding proteins rich in mitochondrial function, such as PGC1α, which can affect mitochondrial activity [21]. PGC1α acts as a key regulator of mitochondrial biogenesis and it has been shown that PGC1α degradation inhibits mitochondrial biogenesis [26, 27]. Activated PGC1α leads to increased expression levels of NRF1 and NRF2 [26]. NRF2 affects mitochondrial membrane potential, which is a universal indicator of mitochondrial health and cellular metabolism [21]. Additionally, Nrf2 deficiency results in impaired mitochondrial function (mitochondrial depolarization, reduced ATP levels) [28]. SIRT1 is another marker of mitochondrial function and activation of the SIRT1/PGC-1α axis prevents aberrant mitochondrial fission [29]. In the present study, YBX2 KD decreased mitochondrial activity and expression levels of NRF1, NRF2, PGC1α, and SIRT1, indicating that YBX2 KD impaired mitochondrial biogenesis.
Additionally, mitochondrial dysfunction can affect ZGA during early embryonic development [3, 22]. Furthermore, mitochondrial metabolism has a profound effect on histone modifications required for gene expression [30]. Therefore, impairment of mitochondrial function caused by YBX2 KD may also affect ZGA. During ZGA, the gradual degradation of most maternal factors is essential for the proper progression of embryonic development. Failure to degrade these maternal factors can result in developmental issues within the embryo [31]. In the present study, YBX2 KD resulted in a significant increase in maternal mRNA expression levels and a decrease in protein expression levels, suggesting that maternal gene degradation and translation were disrupted. Moreover, ZSCAN4 and DPPA2 are ZGA markers [32,33]. Studies have reported that either Zscan4 KD or its overexpression impairs early embryonic development [34]. DPPA2 and DPPA4 are positive regulators of 2C-like cells and ZGA gene transcription [32]. Upon YBX2 KD, we observed a significant reduction in the mRNA levels of both ZSCAN4 and DPPA2, suggesting that YBX2 can affect ZGA. Regulation of ZGA is influenced by histone modifications, including H3K9ac and H3K9me3. H3K9me3 is associated with constitutive heterochromatin formation and repression of genes transcription [35]. H3K9ac is associated with gene transcription activation. In general, gene activation implies that chromatin shifts to an open state; in gene-regulated regions, active markers are enriched and repressive markers disappear [4,36,37].YBX2 KD increased H3K9me3 levels, whereas H3K9ac levels were reduced, indicating the repression of gene transcription. Taken together, these results indicated that YBX2 regulates the ZGA process during embryonic development in pigs.
In conclusion, the current study showed that YBX2 KD decreased mitochondrial biogenesis, reduced transcriptional activity, and caused abnormal histone modifications to impair ZGA. Our research provides insights into the mechanism through which YBX2 modulates the developmental capacity of embryos in vitro, and opens avenues for developing strategies to enhance embryonic viability and potentially improve assisted reproductive technologies.
















