Background
The Inner Mongolian cashmere goat is a Chinese indigenous breed characterized as a double-coated species. The outer coat consists of coarse guard hairs and the undercoat is the soft and precious cashmere. Two kinds of hair follicles which known as primary hair follicles and secondary hair follicles existed in the skin of the Inner Mongolian cashmere goat. Cashmere, which is derived from the secondary hair follicles, has smaller diameters than wool fibers produced by the primary hair follicles. Primary hair follicles and secondary hair follicles form at different periods and play different roles in the development of hair. In mice, the primary hair follicles arose in utero from embryonic day (E) 12.5 and the secondary follicles started to develop until E17 [1, 2]. In goat embryos, the precursor primary follicles were observed in head, neck, shoulder, and belly at E45. The hair follicles gradually formed during 55E to 65E and developed into the mature primary follicles at E135 [3]. The morphogenesis and development of the secondary follicles were similar to those of the primary follicles. The secondary follicles grew from E65 to E75 and then extended to skin surface. The complete structure of the secondary follicle was formed at E135 in the embryos of Chinese cashmere goats [4]. Furthermore, the periodic growth of the secondary follicles also presented in a breed-specific manner [5]. All primary follicles but few secondary follicles were mature at birth and the number of secondary follicles increased 10-fold in the 57 days after birth. The number of primary follicles showed a tendency to decline between 57 and 107 days of age in Australian cashmere goats [6]. Like the hair follicle cycling in other mammals, the growth of cashmere in goats was also tightly programmed by the three synchronized interchanging stages, anagen (growth phase), catagen (regression phase) and telogen (resting phase) throughout postnatal life [7, 8].
Some genes involved in the growth and development of hair follicles in cashmere goats have been identified, such as KAPs [9, 10], BMP [11], Prolactin [12], and Keratin [13, 14]. Furthermore, in mammals, the thyroid hormones (TH) have multi-functions in many important physiological processes including the normal growth, development, differentiation and metabolism. Recently, new insights into TH biological function have been obtained from animal studies involving in epidermis, dermis and hair cycling including anagen prolongation and stimulation of both hair matrix keratinocyte proliferation and hair pigmentation [15, 16]. The metabolism of TH is related to deiodinase, which is also regulated cashmere growth by altering its activity in skin tissue [17, 18]. TH action could be mediated through the thyroid hormone receptor (TR), which is part of the nuclear hormone receptor superfamily and bound to TH in three patterns identified in skin [19, 20]. TR interacts with the hairless gene product, a transcription factor required for hair growth. TR has been detected in epidermal keratinocytes, skin fibroblasts and a number of cell types that made up the hair follicles. In addition, the retinoic acid (RA) is essential for the development and maintenance of hair cycling [21]. The cellular RA-binding protein type II (CRABPII) is involved in RA synthesis pathway, which could shuttle RA to its receptor in nucleus and increase its transcriptional efficiency [22]. Therefore, the dynamic expression of CRABPII mRNA could affect the concentration of RA, which acts on the formation of cashmere though regulating sebaceous gland. During hair cycle, the expression pattern of the RA synthesis and signaling including Crbp, Dhrs9, Aldh1a1, Aldh1a2, Aldh1a3 and Crabp2 defined in rodents only [23].
Based on the genetic studies in humans and rodents, TRα and CRABPII acted important roles in driving the progression of the hair cycle. We postulate that these two genes might have functions during cashmere formation in goat. So in this study, we described the characteristics of TRα and CRABPII genes in the Inner Mongolian cashmere goat and identified their expression patterns in skin tissue during the middle late embryonic stages (E70 to E130).
Materials and methods
The Inner Mongolia cashmere goat is a traditional outstanding breed, which is famous for its excellent cashmere performance and strong adaptation to the semi-desert and desert steppes. The tested individuals were selected from the Aerbasi White Cashmere Goat Breeding Farm in Inner Mongolia Province, China. Twenty-one embryos (three samples at each stage) were randomly collected and any lineage was avoided during the sampling process. Skin samples (approximately 1 cm2 for each individual) were collected from right mid-side of embryos at seven different embryonic days (E70, E75, E80, E90, E100, E120 and E130). Tissue was frozen in liquid nitrogen and stored at −80 °C for further analysis. All the experimental procedures for this experiment were conducted under a protocol approved by the Institutional Animal Care and Use Committee in the College of Animal Science and Technology, Sichuan Agricultural University, China.
The frozen skin tissues were ground using mortars in liquid nitrogen and the total RNA was isolated by Trizol reagent (Invitrogen, Carlsbad CA, USA) according to the manufacturer's protocols. The concentration and quality of the total RNA were further assessed using the NanoDrop spectrophotometer (Bio-Rad, Benicia, USA). The RNase-free DNase I (Promega, Madison, USA) was used to digest genomic DNA. The first-strand cDNA was synthesized using the M-MLV reverse transcriptase kit (Promega, Madison, USA) with oligo (dT) primer.
Three primer pairs were designed to amplify the caprine TRα and CRABPII genes according to their conserved regions of homologies from human, mouse, cattle, sheep and pig (Table 1). PCR was carried out in a 25 μL reaction mixture containing 2 μL first-strand cDNA, 50 mM KCl, 10 mM Tris–HCl (pH 8.3), 0.5 mM MgCl2, 10 pmol each primer, 150 μM dNTPs and 1 unit Taq polymerase (TaKaRa, Dalian, China). The cycling condition included an initial denaturation step at 95 °C for 5 min, 38 cycles of at 94 °C for 30 s, annealing temperature for 30 s and extension at 72 °C for 45 s, and a final extension at 72 °C for 7 min in a PTC-100 PCR thermocycler (MJ Research, Inc., Watertown, MA). PCR products were ligated with the pMD19-T vector (TaKaRa, Dalian, China) after purification, and sequenced by Invitrogen Biotech Co. Ltd. (Shanghai, China).
The quantitative PCR (qRT-PCR) was carried out in an iCycler iQ Real-Time PCR Detection System (Bio-Rad, Benicia, USA) with a total volume of 20 μL containing 10 μL 2 × SYBR Premix Ex Taq II, 0.6 μL primers (10 μM) and 1 μL diluted cDNA. PCR reaction was as follows: a 95 °C denaturation for 30 s, followed by 40 cycles of 94 °C for 15 s, annealing temperature for 30 s, and 72 °C for 30 s. A melting program ranging from 55 °C to 95 °C with a heating rate of 0.5 °C/10 s was carried out to create the melt curves. Reactions were performed in triplicate and negative control was also performed in parallel.
In the present study, three internal control genes (ACTB, GAPDH and TOP2B, Table 1) were selected to normalize the expression levels of TRα and CRABPII mRNAs. To accurate expression profiling of target genes, the geometric mean of multiple carefully selected housekeeping genes was validated as an accurate normalization factor [24]. The relative gene expression was calculated with the 2-ΔΔCt method [25]. Data were presented as mean ± SE. Comparisons between groups were analyzed via GLM (General Linear Model) for experiments with more than 2 subgroups. The significance level was P < 0.05.
Results and discussion
A 1,309-bp fragment of TRα was assembled by the two overlapped sequences of TRα-1 F/1R and TRα-2 F/2R with an open reading frame (ORF) extending from nucleotide positions 21 to 1,253 (with reference to the translational start codon of ATG), which encoded a protein with 410 amino acids (Accession No. KF589923). The obtained sequence of CRABPII mRNA was 563 bp in length with an ORF of 417 bp encoding 138 amino acids (Accession No. KF589924). The blast results revealed that both of TRα and CRABPII were quite conserved among species (Fig. 1, Additional file 1: Figure S1 and Additional file 2: Figure S2). The sequence similarity ranged from 88 % to 100 % (Additional file 3: Table S1). The coding sequence of the caprine TRα gene shows a high similarity with the sequences in other mammals, sharing 99 % identity with sheep (NM_001100919) and cattle (NM_001046329). The goat CRABPII shows 88 % identity with mice (NM_007759) and 98 % identity with cattle (NM_001008670).
The nucleotide sequences were aligned by the Cluster W method included in the program BioEdit version 7.2.5 [26]. The phylogenetic analysis was constructed using the program MEGA 4.1 [27], with a Kimura 2-parameter model and a bootstrap test (1000 replications). The phylogenetic tree revealed that the goat TR grouped with sheep, and then clustered with cattle, pig, human, mice and chicken subsequently (Fig. 2a). The phylogenetic tree of CRABPII gene showed a similar clustering with differences in sort of branch-length groups (Fig. 2b). The Minimum Evolution, Maximum Parsimony and UPGMA trees revealed the same clustering groups as presented by the NJ trees (data not shown).
To better understand the prenatal dynamical expressions of TRα and CRABPII in the skin tissue of cashmere goats, the qRT-PCR array was performed in the middle late embryonic stages (E70 to E130). As shown in Fig. 3, both of TRα and CRABPII mRNAs were detectable in all the tested time points. However, no significant difference of TRα gene expression was detected during the middle late development of goat embryos. The mRNA of TRα was expressed at E70 with relatively high level and mildly decreased in the following three stages (E75, E80, and E90), and then increased at E100 and reduced to the lowest level at E120, subsequently. The highest expression of TRα gene was observed in the last stage (E130, P > 0.05). The previous studies has reported that the secondary follicles grew from E65 to E75 and then extended to skin surface. The complete structure of the secondary follicle was formed at E135 in Chinese cashmere goats [4, 28, 29]. Synchronously coupled with the early formation and growth of cashmere, the mRNA expression of TRα gene was up-regulated indicating that TRα could play a role in the time-course growth of goat cashmere.
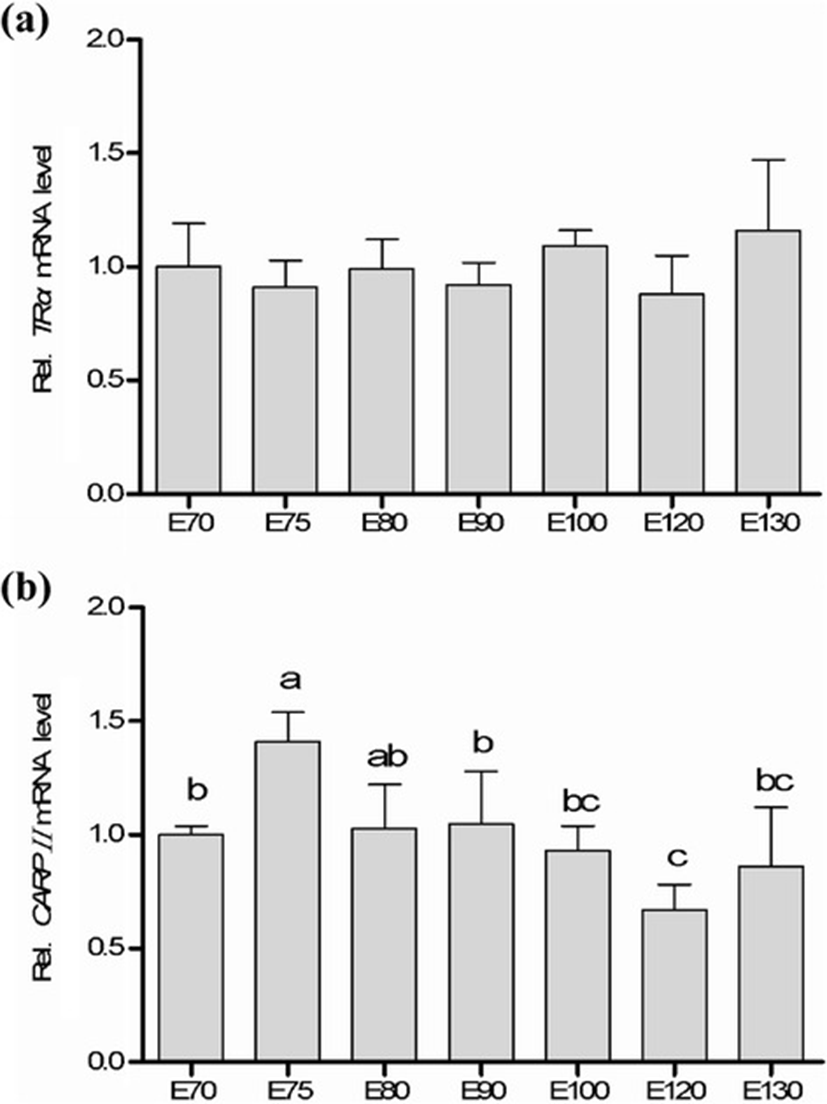
The expression pattern of CRABPII mRNA showed an “up-down-up” trend, which revealed a significantly highest expression at E75 (P < 0.05), and was down-regulated during E80 to E120 (P < 0.05) then increased again at E130. In embryonic development of hair follicles, the glandula sebacea cells were observed in the skin tissue from cashmere goat fetus at E85 [30]. The glandula sebacea formed at E90 and accelerated the growth of primary hair follicles. However, the physiologic difference between primary and secondary follicles was that no glandula sebacea was found in secondary hair follicles. The second hair follicles grew retard and partially matured at E130. The mRNA expression of CRABPII at E90 was lower than that at E80 when no glandula sebacea was formed. The CRABPII gene could regulate the early development of glandula sebacea though modifying the concentration of RA. The mRNA of CRABPII gene at E100 expressed significantly higher than that at E120, which led more RA transported into nucleus and bound to its receptor, and proposed to boost the growth of glandula sebacea. In humans, the concentration of RA in cells could increase the mRNA expression of CRABPII in skin [31, 32].
In this study, we characterized the caprine TRα and CRABPII genes and quantified their mRNA expressions during the formation of secondary hair follicles in the middle late embryonic periods. Our study will enrich the knowledge of goat TRα and CRABPII genes and provide the foundation for further insight into their functions on cashmere growth.
Conclusions
Cashmere wool is the very valuable production obtained from goats. It is very important to investigate the expressions of key functional genes associated with cashmere growth during the prenatal and process-oriented periods (from anagen to catagen and finally telogen). Taken together, our results profiled the expressions of TRα and CRABPII genes associated with prenatal development of goat hair follicle.

















