INTRODUCTION
A number of Bacillus strains reported to degrade non-digestible carbohydrates, including cellulose and hemicellulose, have been used as feed additives to increase feed efficiency [1,2]. Bacillus amyloliquefaciens ATC6 was originally isolated from pig feces, and produces acidic cellulase [3]. Here we sequenced the complete genome of B. amyloliquefaciens ATC6. B. amyloliquefaciens ATC6 was grown in Nutrient Broth (Difco, Franklin Lakes, NJ, USA) at 37°C for 12 h. The genomic DNA was prepared as described previously [4], and the genome quality was checked using a spectrophotometer (UV-1601PC, Shimadzu, Kyoto, Japan). The genomic DNA of B. amyloliquefaciens ATC6 was sequenced using the PacBio RSII platform (ver. 2.0; Pacific Biosciences, CA, USA) at Macrogen. (Korea). All generated reads were de novo assembled using RS HGAP Assembly (ver. 3.0) [5]. Functional annotation was performed using the InterProScan (ver. 5.30–69.0) (http://www.ebi.ac.uk/interpro/), GO (http://geneontology.org/page/go-database), BLAST (ver. 2.6.0+) (http://geneontology.org/page/go-database) with UniProt (ver. 2018_06) (http://www.uniprot.org/), and EggNOG (ver. 4.5) (http://eggnogdb.embl.de/#/app/home) databases, and the genome was submitted to GenBank (https://www.ncbi.nlm.nih.gov/) (No. PRJNA639017).
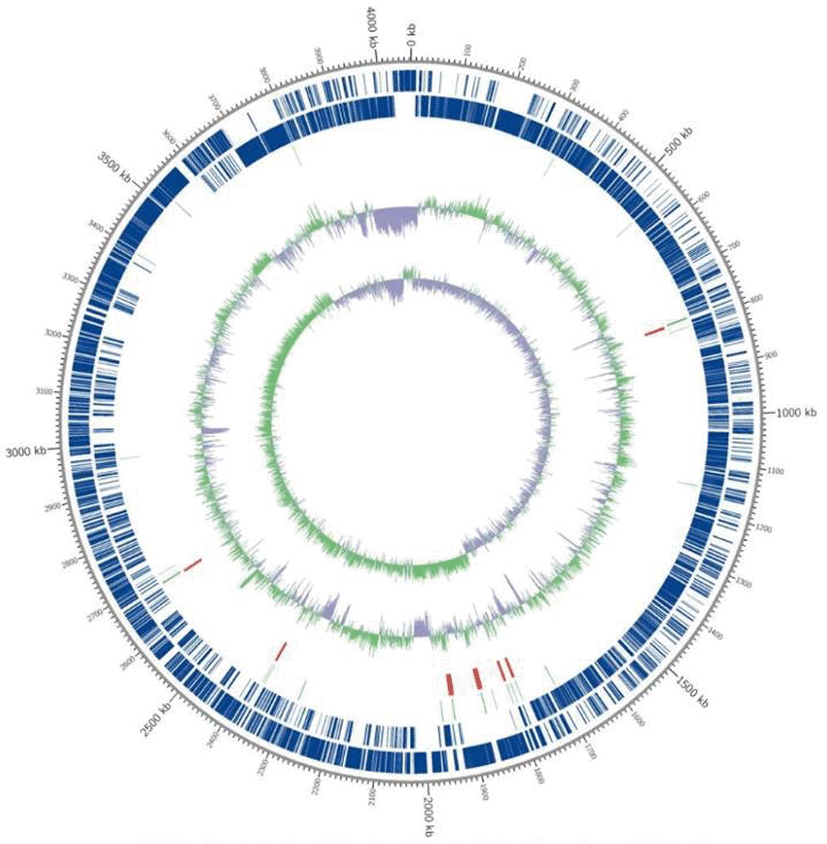
The complete genome of strain ATC6 was 4,062,817 bp in length with a guanine-cytosine (GC) content of 46.27% (Table 1). No plasmid was detected (Fig. 1). The chromosome contained a total of 3,913 protein-coding sequences, 27 ribosomal RNAs, and 86 transfer RNAs that were annotated (Table 1). Analysis of the genomic sequences revealed that ATC6 possessed genes involved in cellulose degradation, as shown in our previous report [3]. This genome also contained genes associated with the degradation of other non-digestible carbohydrates, such as xylan and mannan. The genomic information of B. amyloliquefaciens ATC6 will help clarify the biodegradation mechanism of non-digestible carbohydrates, which could increase animal feed efficiency.
| Attribute | Value |
|---|---|
| Genome size (base pair) | 4,062,817 |
| GC content (%) | 46.27 |
| No. of contigs | 1 |
| Total genes | 4,026 |
| Protein-coding genes | 3,913 |
| tRNAs | 86 |
| rRNAs | 27 |