INTRODUCTION
Infectious calf diarrhea is one of the most significant diseases of neonatal calves. It has affected the morbidity and mortality of neonatal calves and their growth performances and has caused worldwide economic loss [1]. Even though various methods have been designed to treat calf diarrhea, prevention is still the best approach to reduce the disease, and monitoring for pathogens is one of the most important preventive actions [2]. Many researchers and reports worldwide have attempted to determine the prevalence of infectious pathogens in calf diarrhea [3–5]. Major pathogens causing calf diarrhea in these reports were: viruses (bovine coronavirus [BCV], bovine rotavirus group A [BRV], and bovine viral diarrhea virus [BVDV]), bacteria (Escherichia coli K99 and Salmonella spp.), and protozoa (Cryptosporidium parvum and Eimeria spp.). Some of the agents are known to be detected not only in diarrheic calves but also in normal calves.
In Korea, like other countries, calf diarrhea has had a serious impact on calf death. According to previous studies, 68.7% of calf deaths in Korean native beef calves and 53.4% in dairy calves were caused by digestive diseases [6–7]. Additionally, there have been several recent reports investigating pathogens that cause calf diarrhea [8–10]. However, most of them have been focused on specific pathogens from calf feces. As calf diarrhea can be caused by a variety of pathogens, it is necessary to simultaneously analyze different kinds of pathogens.
This study was performed to investigate the distribution of causative agents of calf diarrhea in Korean native beef calves aged less than 60 days in various regions of Korea and to discern their association with diarrhea.
MATERIALS AND METHODS
In this study, calves up to 60 days of age in 10 local Korean indigenous cattle farms in different areas of Korea (Yeongju, Samnye, Asan, Gimje, Mungyeong, Wanju, Heongseong, Sancheong, Iksan, Sangju) were selected for feces collection from 2016–2017. Feces were obtained by digital rectal palpation from the calves. All feces were scored as 0 to 3 using the scoring system included in the calf health scoring guide created by the University of Wisconsin-Madison School of Veterinary Medicine [11] and stored in 50 mL specimen bottles (SPL Life Sciences, Pocheon, Korea) at 4°C until they were transported to the laboratory. All feces scored at 2 and 3 were categorized as diarrhea.
All samples were examined for 7 pathogens (BCV, BRV, BVDV, C. parvum, Eimeria spp., E. coli K99, Salmonella spp.). Each feces sample was divided into two tubes and treated differently depending on the target agent, according to previously reported methods [8–12]. Briefly, to detect the 6 pathogens causing calf diarrhea (BCV, BRV, BVDV, C. parvum, E. coli K99, Salmonella spp.), fecal samples were suspended in 0.01 M phosphate-buffered saline to make 30% fecal homogenates and centrifuged for 1 min at 100×g. A supernatant was used to extract the total nucleic acid using MagMAXTM Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). All extracts were stored at −70°C until real-time polymerase chain reaction (PCR) was performed. Real-time PCR was performed with the Path-IDTM Multiplex One-Step RT-PCR kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s recommended protocols in a 25 uL reaction volume using 8 ul of extracted template and 17 uL of the reaction mixture. Two types of real-time PCR were performed using specific primer sets for each pathogen in Table 1: one for the 3 viruses (BCV, BRV, BVDV) and the other for the bacteria and protozoa (C. parvum, E. coli K99, Salmonella spp.). Equal volumes of primers and probes were mixed for each target agent and the final concentration of each primer and probe was 0.2 uM. Real-time PCR was performed using ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Cycling conditions of real-time PCR were as follows: (a) reverse transcription (RT) for 30 min at 45°C; (b) activation of DNA polymerase for 10 min at 95°C; (c) 40 cycles of denaturation at 94°C for 15 sec and annealing/extension at 60°C for 60 sec. RT step was performed only for viruses. After a 40 cycles reaction, samples with cycle threshold value less than 35 for targets were considered positive. To detect Eimeria spp., all fecal samples were suspended in a solution of 2.5% potassium dichromate and then transported to the laboratory. In the laboratory, fecal samples were analyzed to detect oocysts using the floatation methods with Sheather’s solution (saturated sugar solution; specific gravity 1.28) and examined microscopically (×400 magnification) based on the morphological features of the oocysts of the Eimeria spp.
| Type | Microbial agents | PCR primers, probes and conditions | Primer sequences (5’ - 3’) | Reference | |||
|---|---|---|---|---|---|---|---|
| Reverse transcription (°C/min) | Activation of DNA polymerase (°C/min) | Denaturation (°C/min) | Annealing/extension (°C/min) | ||||
| Viruses (PCR type 1) | Bovine viral diarrhea virus | BVD-F | GGG NAG TCG TCA RTG GTT CG | [23] | |||
| BVD-R | GTG CCA TGT ACA GCA GAG WTT TT | ||||||
| BVD-Probe (CY5/BHQ2) | CCA YGT GGA CGA GGG CAY GC | ||||||
| Bovine coronavirus | BCV-F | CTA GTA ACC AGG CTG ATG TCA ATA CC | [12] | ||||
| BCV-R | GGC GGA AAC CTA GTC GGA ATA | ||||||
| BCV-Probe (FAM/MGB) | CGC CTG ACA TTC TCG ATC | ||||||
| Bovine rotavirus | BRV-F | TCA ACA TGG ATG TCC TGT ATT CCT | [24] | ||||
| BRV-R | TCC CCC AGT TTG GAA TTC ATT | ||||||
| BRV-Probe (VIC/MGB) | TCA AAA ACT CTT AAA GAT GCA AG | ||||||
| Conditions | 45/10 | 95/10 | 95/0.25 | 60/1 | |||
| Bacteria/parasites (PCR type 2) | Escherichia coli K99 | K99-F | GCT ATT AGT GGT CAT GGC ACT GTA G | [25] | |||
| K99-R | TTT GTT TTC GCT AGG CAG TCA TTA | ||||||
| K99-Probe (FAM/BHQ1) | ATT TTA AAC TAA AAC CAG CGC CCG GCA | ||||||
| Cryptosporidium parvum | Cryptosporidium parvum-F | CAA ATT GAT ACC GTT TGT CCT TCT GT | [26] | ||||
| Cryptosporidium parvum-R | GGC ATG TCG ATT CTA ATT CAG CT | ||||||
| Cryptosporidium parvum-Probe (JOE/BHQ1) | TGC CAT ACA TTG TTG TCC TGA CAA ATT GAA | ||||||
| Salmonella species | Salmonella-F | GGG NAG TCG TCA RTG GTT CG | [27] | ||||
| Salmonella-R | GTG CCA TGT ACA GCA GAG WTT TT | ||||||
| Salmonella-Probe (CY5/BHQ2) | CCA YGT GGA CGA GGG CAY GC | ||||||
| Conditions | N/A | 95/10 | 95/0.25 | 60/1 | |||
The PCR results for each pathogen were recorded as positive or negative and categorized based on diarrhea status and age group. Age group was divided into three age group 1 (1 d–10 d), age group 2 (11 d–30 d,), and age group 3 (31 d–60 d). All statistical methods (The χ2, Fischer’s exact tests, and linear by linear association) were performed by SPSS v. 25.0 (IBM, Armonk, NY, USA). All graphical works were performed by GraphPad Prism 6 software (GraphPad, San Diego, CA, USA).
RESULTS
Fecal samples collected from 544 Korean native beef calves on 10 local Korean indigenous cattle farms were described in Table 2. According to our results, diarrhea was not significantly associated with age group. The presence of pathogens in non-diarrheic calves was compared to that in diarrheic calves. Of 340 non-diarrheic calves, 213 calves (62.6%) were negative and 127 calves (37.4%) were positive for the pathogens examined. Alternatively, of 204 diarrheic calves, 101 calves (49.5%) were negative and 103 calves (50.5%) were positive for the pathogens. The presence of pathogens was significantly associated with diarrhea (odds ratio = 1.71, 95% confidence interval = 1.203–2.431, p < 0.01). And also there was a significant linear trend when comparing fecal scores and the number of detected agents (Fig. 1, p < 0.001).
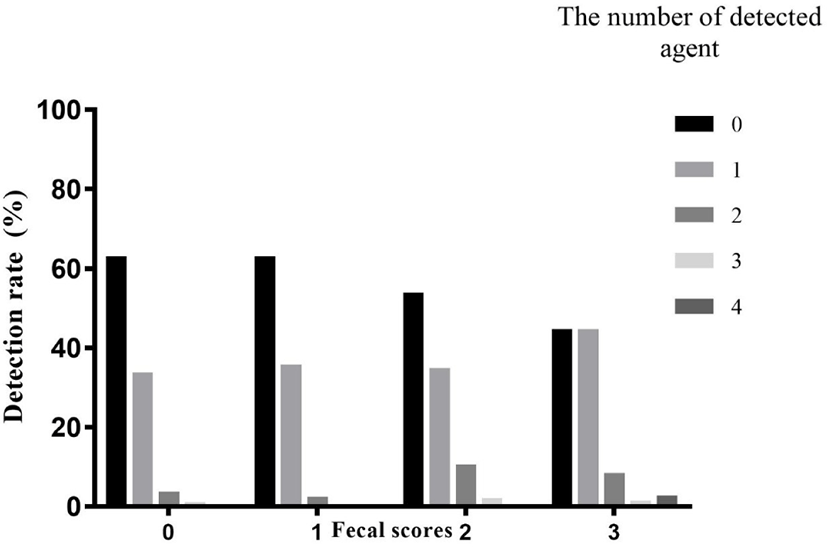
The detection rate of the 7 pathogens in the normal feces and diarrheic feces is described in Table 3. Eimeria spp. (27.4%) was the most detected pathogen in overall samples, followed by BRV (8.8%), BCV (8.5%), C. parvum (4.4%), BVDV (0.7%), and E. coli K99 (0.2%). There was no Salmonella spp. in any our samples. In the diarrheic samples, Eimeria spp. (31.4%) was detected most often, followed by BRV (15.2%), BCV (10.3%), C. parvum (8.3%), and E. coli K99 (0.5%). No BVDV or Salmonella spp. was detected. C. parvum (p = 0.001) and BRV (p < 0.001) had a significantly higher presence in diarrheic calves than in non-diarrheic calves.
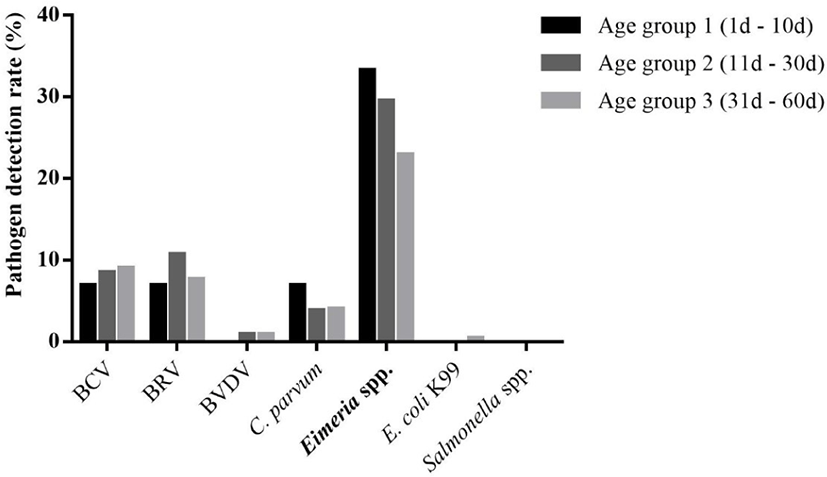
The detection rate of each pathogen according to age group was also compared (Fig. 2). Eimeria spp. was detected 33.3% (29/87), 29.5% (69/234), and 22.9% (51/223) in age group 1, 2, and 3, respectively. There was a significant linear trend between the detection rate of Eimeria spp. and the age group (p < 0.05). BRV was detected 6.9% (6/87), 10.7% (25/234), and 7.6% (17/223) in age group 1, 2, and 3, respectively. There was no significant linear trend between the detection rate of BRV and the age group. BCV was detected 6.9% (6/87), 8.5% (20/234), and 9.0% (20/223) in age group 1, 2, and 3, respectively. There was no significant linear trend between the detection rate of BCV and the age group. C. parvum was detected 6.9% (6/87), 3.8% (9/234), and 4.0% (9/223) in age group 1, 2, and 3, respectively. There was no significant linear trend between the detection rate of C. parvum and the age group. BVDV was detected 0% (0/87), 0.9% (2/234), and 0.9% (2/223) in age group 1, 2, and 3, respectively. There was no significant linear trend between the detection rate of BVDV and the age group. E. coli K99 was detected 0% (0/87), 0% (0/234), and 0.4% (1/223) in age group 1, 2, and 3, respectively. There was no significant linear trend between the detection rate of E. coli K99 and the age group.
DISCUSSION
In this study, the prevalence of the 7 pathogens in normal and diarrheic calves and the association between the pathogens causing calf diarrhea and the age and fecal status of 544 Korean native beef calves were demonstrated. As expected, diarrheic calves (50.5%) showed a significantly higher positive rate of pathogens than normal calves (37.4%), and the fecal consistency had a linear association with the number of detected pathogens, consistent with findings from other countries [13]. This suggested that pathogens were the one of the primary factors related to diarrhea in Korean native beef calves.
Three viruses (BRV, BCV, and BVDV) were detected in Korean native beef calves. BRV was detected 15.2% in Korean native beef calves and significantly related to diarrhea (p < 0.001). In other reports in Korea, BRV was detected in 34.8% from diarrhea feces in Korean native calves [14], which might be come from the difference of regions, research periods, and methodology. However, these results including previous reports demonstrate that rotavirus is an important pathogen that can negatively affect the health of calves, consistent with that of earlier reports [13–15]. BCV was detected in non-diarrheic and diarrheic calves and there was no significant difference. Even though BCV is known as one of the main pathogens associated with calf diarrhea, this result that BCV were detected in normal feces was similar to that seen in earlier reports [3–16]. BVDV was detected in only 4 calves and all of them were in the non-diarrheic group. The detection rate of BVDV in this study was less than that in previous research [17]. This result might come from the type of samples. Feces were used to detect BVDV in this research, however, ear notch, skin fold biopsies, and nasal swabs showed reliable results for the detection of BVDV than rectal swab [18].
Even though the detection rate of C. parvum was lower than that for Eimeria spp. and BCV, C. parvum was found at a significantly higher rate in diarrheic calves than in normal feces, similar to BRV. There have been many reports emphasizing the effects of C. parvum infection in calf diarrhea in other countries [3–13,19]. Because there is no worldwide commercial vaccine for C. parvum, maintaining good herd sanitation and keeping sick calves away from non-diarrheic calves are important in preventing C. parvum infections.
Two bacteria (E. coli K99 and Salmonella spp.) were selected in this research. There was only one calf positive for E. coli K99 in this research. This result was consistent with that of other reports in Korea that no E. coli strain expressing K99 was detected in isolated samples from cattle farms [20]. Salmonella spp. occurring calf diarrhea was not detected in this research. However, since Salmonella infection in other livestock and human have been reported in Korea [21–22], it is necessary to conduct ongoing monitoring of Salmonella infection in Korean beef calves.
In this study, Eimeria spp. was the most detected pathogen of the 7 examined pathogens and this detection rate was similar to that in other reports from Korea [8]. However, no significant difference was shown between non-diarrheic calves and diarrheic calves. Because Eimeria spp. was also detected frequently in the feces of non-diarrheic calves [23], this result was conceivable. The amount of oocyte secretion was not investigated in this research, but the amount of oocytes excretion of Eimeria spp. is known to be strongly correlated with diarrhea, and thus, further research should investigate the correlation between diarrhea in Korean native beef calves and the amount of Eimeria spp. excreted.
In comparing the age groups among calves to the pathogens detected, only Eimeria spp. showed a linear association to the age groups (Fig. 2). The prevalence of Eimeria infections in normal calves decreased as the age increased (p < 0.01, linear trend), while in diarrheic calves, the prevalence was stable even as the age increased (Table 4). According to this result, ongoing investigations of the amount of Eimeria spp. infection are important in predicting the pattern of calf diarrhea by Eimeria speices.
In conclusion, six of seven pathogens were detected in samples, but only C. parvum and bovine rotavirus were found at significantly higher rates in diarrheic feces than in non-diarrheic feces and Eimeria spp. showed a significant linear trend between the detection rate of the pathogen and the age groups.
















