RESEARCH ARTICLE
Complete genome sequence of Bacillus coagulans CACC834 isolated from canine
Jung-Ae Kim1,2
,
Dae-Hyuk Kim1,3
,
Yangseon Kim1,*
Author Information & Copyright ▼
1Department of Research and Development, Center for Industrialization of Agricultural and Livestock Microorganisms, Jeongeup 56212, Korea
2Department of Bioactive Material Science, Jeonbuk National University, Jeonju 54896, Korea
3Department of Molecular Biology, Department of Bioactive Material Science, Institute for Molecular Biology and Genetics, Jeonbuk National University, Jeonju 54896, Korea
*Corresponding author: Yangseon Kim, Department of Research and Development, Center for Industrialization of Agricultural and Livestock Microorganisms, Jeongeup 56212, Korea., Tel: +82-63-536-6712, E-mail:
yangseon@cialm.or.kr
© Copyright 2021 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Jun 21, 2021; Revised: Sep 15, 2021; Accepted: Sep 25, 2021
Published Online: Nov 30, 2021
Abstract
Bacillus coagulans CACC 834 was isolated from canine feces, and its potential probiotic properties were characterized by functional genome analysis. Whole-genome sequencing of B. coagulans CACC 834 was performed using the PacBio RSII platforms. The complete genome assembly consisted of one circular chromosome (3.1 Mb) with guanine (G) + cytosine (C) content of 47.1%. Annotation revealed 3,181 protein-coding sequences (CDSs), 30 rRNAs, and 83 tRNAs. Gene associated 11% of the genes were involved in replication, recombination, and repair. We also annotated various stress-related, acid resistance, bile salt resistance and adhesion-related domains in this strain, which likely provide support in exerting probiotic action by survival under gastrointestinal tract. These results add to our comprehensive understanding of B. coagulans and suggest potential mammal-related industrial applications.
Keywords: Bacillus coagulans; Canine; Whole-genome sequencing
Bacillus coagulans strains are gram-positive, spore-forming, and produce lactic acid, which possesses the capacity to balance intestinal gut microbiota, ultimately promoting the growth of animals and improving immunity [1,2]. In addition, B. coagulans is resistant to high temperatures because of its probiotic activity, modulates and strengthens the immune system to protect against infections, and minimizes inflammation-related tissue damage [3].
We isolated B. coagulans CACC 834 (KACC 22145) from the feces of a female 3-year-old Boston terrier in Korea. The samples were incubated under anaerobic conditions at 37°C [4].
Whole-genome sequencing was performed using the Pacific Biosciences (PacBio) RS II Single Molecule Real Time (SMRT) platform with a 20 kb SMRTbellTM template library at ChunLab. The reads were assembled using the PacBio SMAR Analysis 2.3.0. [5].
The sequences were annotated using the combined results of the automatic National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) and Rapid Annotations using Subsystems Technology (RAST) [6].
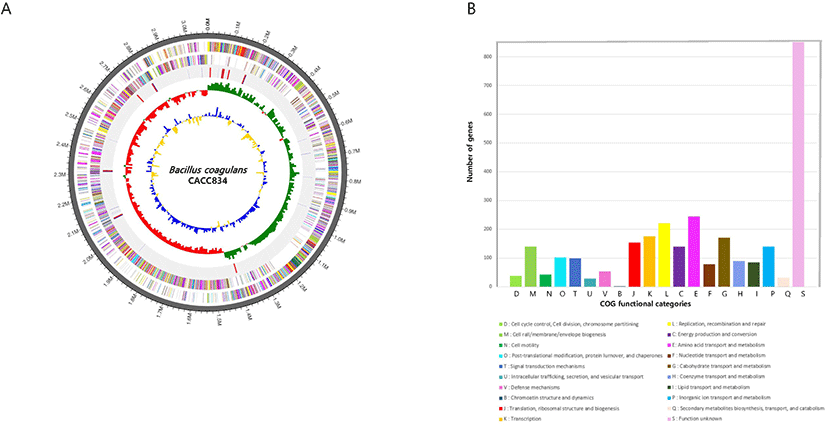
The complete genome of B. coagulans CACC 834 was composed of a 3,077,319 bp circular chromosome with 47.1% guanine (G) + cytosine (C) content. The genome contained 3,181 protein-coding sequences (CDS), 30 rRNAs, and 83 tRNAs (Table 1). The genome features of B. coagulans CACC 834 are summarized in Fig. 1. Among these CDS, 2,909 genes were classified into 20 clusters of orthologous groups (COG) functional categories (Fig. 1B). More than 41% of genes were involved in transport and metabolism, including amino acid, carbohydrate, inorganic ion, nucleotide, coenzyme, and lipid. Interestingly, 11% of the genes were involved in replication, recombination, and repair (Fig. 1B).
Table 1.
Genome overview of Bacillus coagulas CACC834
| Feature |
Values |
| Genome size (bp) |
3,077,319 |
| No. of contigs |
1 |
| GC content |
47.1% |
| Protein-coding genes (CDSs) |
684 |
| rRNA |
30 |
| tRNA |
83 |
| plasmid |
0 |
| Genbank Acession No. |
CP076597 |
Download Excel Table
Fig. 1.
Genome features of Bacillus coagulans CACC834.
(A) Circular genome maps of B. coagulans CACC834 chromosome. Circles from the outside to the center denote rRNA and tRNA gene, reverse strand CDS, forward strand CDS, GC skew, and GC content. (B) Genome number of COG categorie. CDS, protein-coding genes; COG, clusters of orthologous group; G, guanine; C, cytosine.
Download Original Figure
The genome of B. coagulans CACC 834 possessed hsp20, hsp60, dnaK, dnaJ and grpE operons, encoding enzymes and proteins for heat shock. Also, B. coagulans CACC 834 has heat shock induced genes, such as clpB, clpP, mcsB, and repair-related genes, such as recA and uvrABC. The expression of these genes is expected to help promote cell recovery from heat shock by limiting damage caused by stress [7–9]. Furthermore, B. coagulans CACC 834 carried genes known to be involved in lactate synthesis, adhesion, acid resistance, and bile resistance (Table 2).
Table 2.
Predicted genes involved in probiotic potency in B. coagulans CACC834
| Predicted function |
B. coagulans CACC834 |
| Predicted genes |
Start position |
End position |
Length (bp) |
| Lactate synthesis |
ldh
|
1,433,259 |
1,434,242 |
984 |
| Adhesion protein |
EpsD
|
393,741 |
394,838 |
1,098 |
|
|
FliD
|
724,445 |
726,531 |
2,088 |
| Acid resistance-related |
|
|
|
|
| Protection or repair of macromolecules |
dnaK
|
1,737,678 |
1,737,546 |
1,827 |
|
|
dnaJ
|
1,736,410 |
1,737,546 |
1,137 |
|
|
grpE
|
1,739,540 |
1,740,202 |
663 |
|
|
recA
|
125,046 |
1,226,113 |
1,068 |
|
|
uvrABC
|
765,135 |
767,135 |
2,001 |
|
|
clpB
|
108,312 |
110,762 |
2,451 |
|
|
clpP
|
793,689 |
794,279 |
591 |
|
|
mcsB
|
108,312 |
110,762 |
2,451 |
| Fatty acid synthesis |
fabF
|
870,816 |
871,754 |
1,242 |
|
|
fabH
|
871,824 |
873,065 |
939 |
|
|
fadD
|
260,088 |
261,242 |
1,155 |
|
|
fabI
|
903,131 |
903,910 |
780 |
| Acid shock response |
aspS
|
1,804,625 |
180,696 |
1,773 |
| Metabolic rearrangements |
alsD
|
2,318,874 |
2,319,623 |
750 |
| Gylcine betain ABC transport system |
opuCC
|
467,919 |
468,833 |
915 |
| Bile salt resistance |
bass
|
1,090,675 |
1,091,145 |
471 |
Download Excel Table
This study on complete genome sequence of B. coagulans CACC 834 may increase our understanding of the probiotic effects in host healthcare and extend its potential application as an industrial strain.
The complete genome of B. coagulans strain CACC 834 determined in this study has been deposited in the NCBI GenBank database under accession number CP076597.